Using generative AI to investigate medical imagery models and datasets
June 5, 2024
Oran Lang, Software Engineer, Google Research, and Heather Cole-Lewis, Health Equity Clinical Scientist, Google Core
Quick links
Machine learning (ML) has the potential to revolutionize healthcare, from reducing workload and improving efficiency to uncovering novel biomarkers and disease signals. In order to harness these benefits responsibly, researchers employ explainability techniques to understand how ML models make predictions. However, current saliency-based approaches, which highlight important image regions, often fall short of explaining how specific visual changes drive ML decisions. Visualizing these changes (which we call “attributes”) are helpful to interrogate aspects of bias that are not readily apparent via quantitative metrics, such as how datasets were curated, how models were trained, problem formulation, and human-computer interaction. These visualizations can also help researchers understand if these mechanisms might represent novel insights for further investigation.
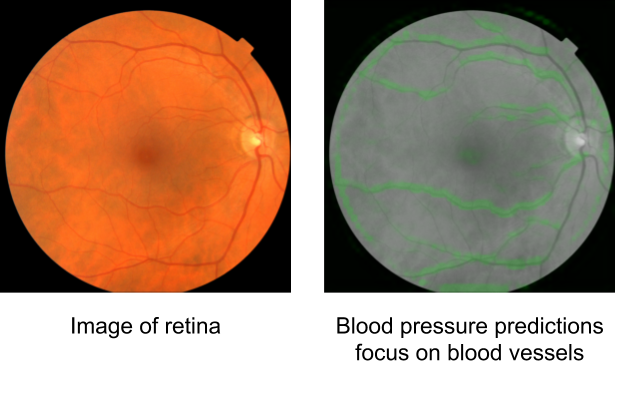
In "Using generative AI to investigate medical imagery models and datasets", published in The Lancet eBioMedicine, we explored the potential of generative models to enhance our understanding of medical imaging ML models. Based upon the previously published StylEx method, which generates visual explanations of classifiers, our goal was to develop a general approach that can be applied broadly in medical imaging research. To test our approach, we selected three imaging modalities (external eye photographs, fundus photos, and chest X-rays [CXRs]) and eight prediction tasks based on recent scientific literature. These include established clinical tasks as “positive controls”, where known attributes contribute to the prediction, and also tasks that clinicians are not trained to perform. For external eye photographs, we examined classifiers that are able to detect signs of diseases from images of the front of the eye. For fundus photos, we examined classifiers that demonstrated surprising results for predicting cardiovascular risk factors. Additionally, for CXRs, we examined abnormality classifiers as well as the surprising capability to predict race.
GenAI framework to investigate medical imagery models and datasets
Our framework operates in four key stages:
- Classifier training:
We train an ML classifier model to perform a specific medical imaging task, such as detecting signs of disease. The model is frozen after this step. If a model of interest is already available, it can be used in a frozen state, without needing to further modify this model. - StylEx training:
Then, we train a StylEx generative model, which includes a StyleGAN-v2–based image generator with two additional losses. The first additional loss is an auto-encoder loss, which teaches the generator to create an output image that resembles the input image. The second loss is a classifier loss, which encourages the classifier probability of the generated image to be the same as that of the input image. Together, these losses enable the generator to produce images that both look realistic and retain the classifier's predictions. - Automatic attribute selection:
We use the StylEx model to automatically generate visual attributes by creating counterfactual visualizations for a set of images. Each counterfactual visualization is based on a real image, but modified using the StylEx generator while changing one attribute at a time (see animations below). Then, the attributes are filtered and ranked to retain the ones that most influence the classifier's decisions. - Expert panel review:
Finally, an interdisciplinary panel of experts, including relevant clinical specialists, social scientists, and more, analyze the identified attributes, interpreting them within their medical and social contexts.
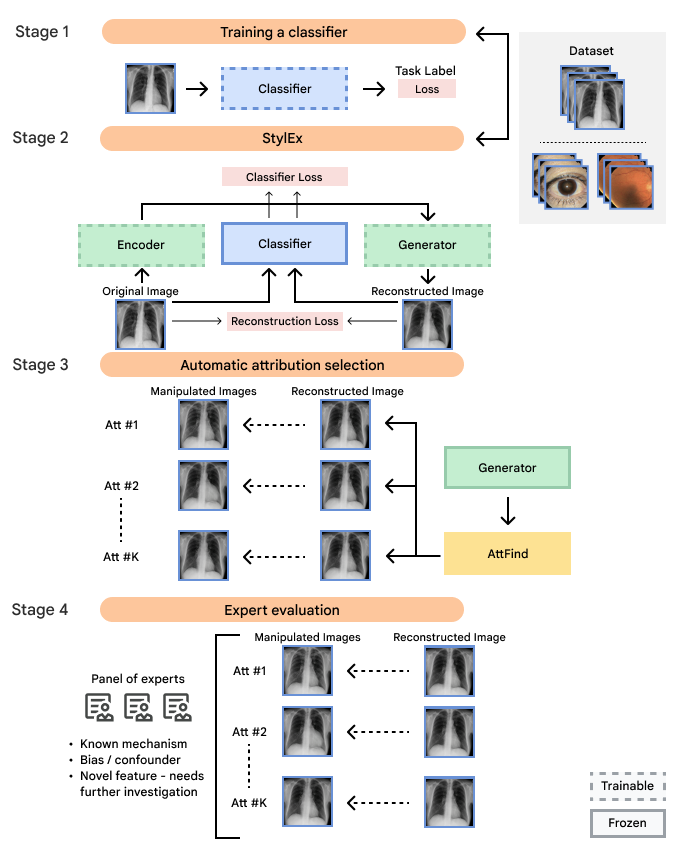
Flowchart of our approach illustrating the four main steps, including (1) developing the ML classifier for a prediction task of interest; (2) developing the generative StylEx ML model to examine the frozen classifier; (3) generating visual attributes using the generative model and extracting the most influential visual attributes; and (4) involving an interdisciplinary panel to examine the features to minimize blind spots in interpretation.
Positive controls
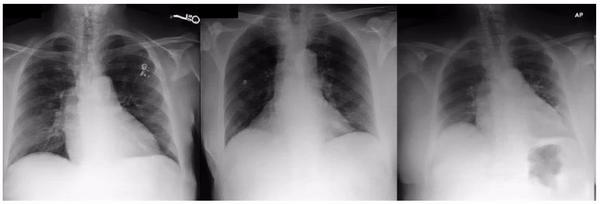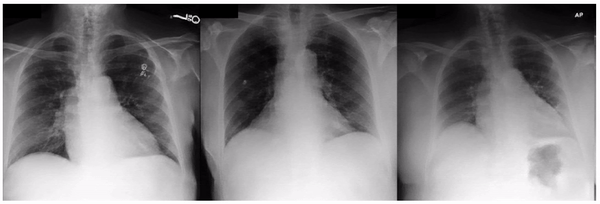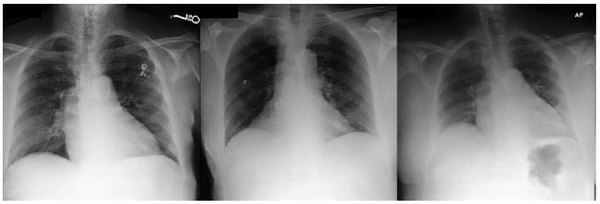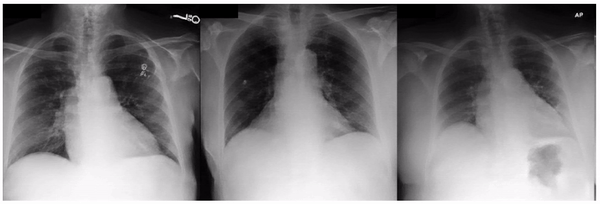
First, to ensure the framework successfully identifies known visual attributes, we examined tasks for each imaging modality, where some known attributes exist (i.e., “positive control” experiments). Indeed, we found that cortical cataract spokes were seen for cataract prediction from external eye photos, retinal vein dilation for smoking status prediction from fundus photos, and left ventricular enlargement for abnormality prediction from CXRs.
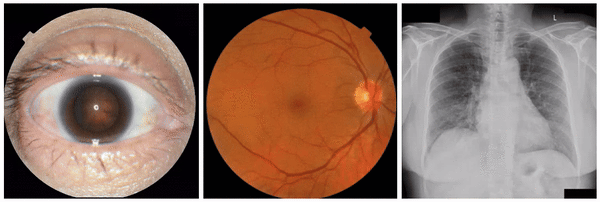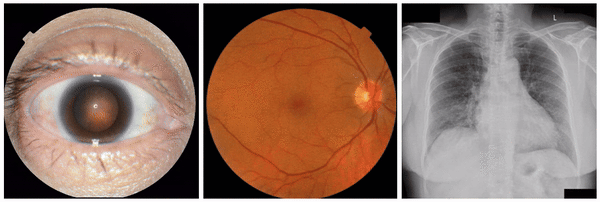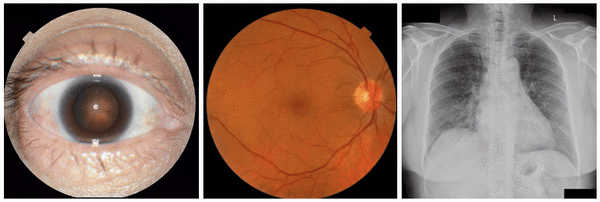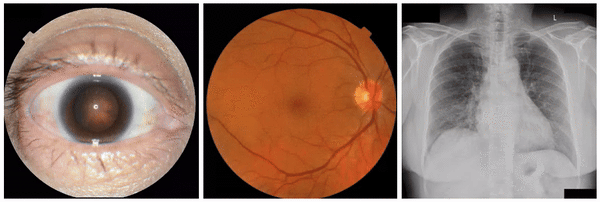
Examples of “known” (positive control) attributes extracted by our approach: a) Spokes for cataract presence, which appear as radial opacities like spokes of a wheel; b) retinal vein dilation for smoking status; c) left ventricular enlargement for abnormal CXR.
Possible novel signals
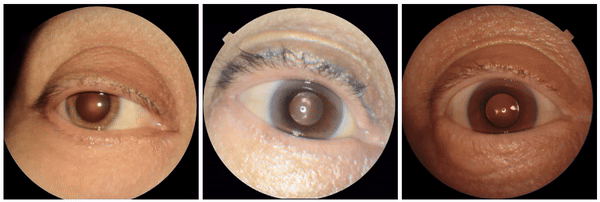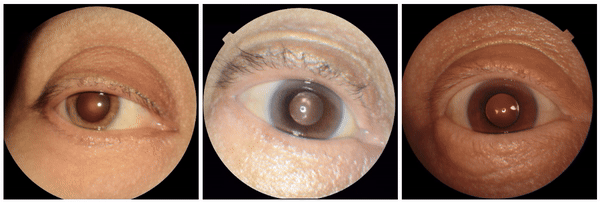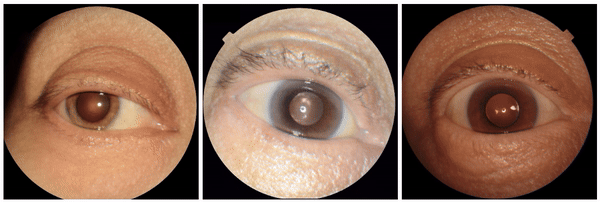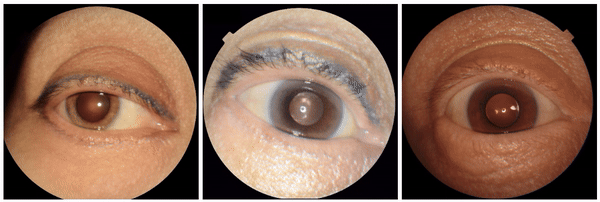
We also discovered visually striking but intriguing associations. One example is an increased eyelid margin pallor correlating with elevated HbA1c levels. This observation aligns with previous research suggesting a link between meibomian gland disease and diabetes, potentially paving the way for further investigation into the underlying mechanisms.
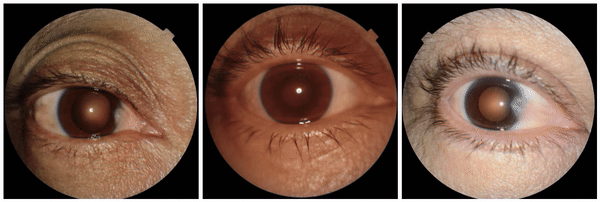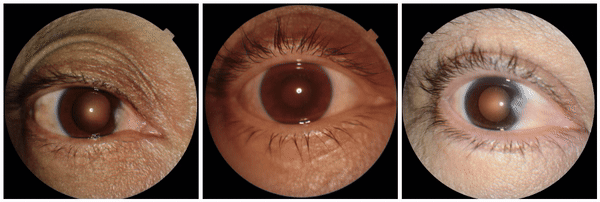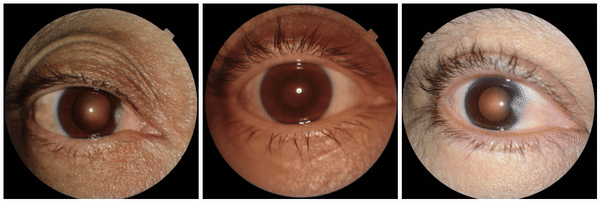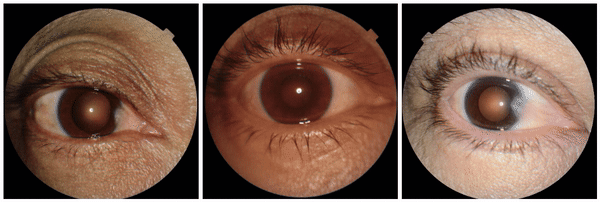
Examples of eyelid margin pallor, an attribute associated with higher predicted HbA1c (a blood sugar measure).
Possible confounders
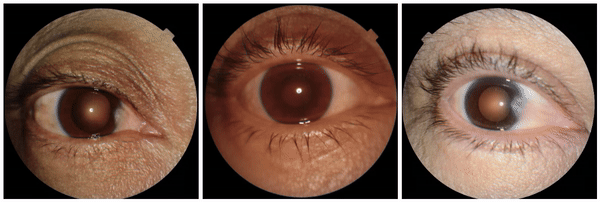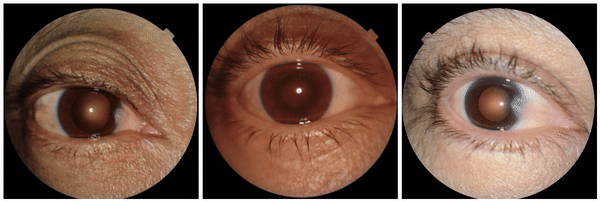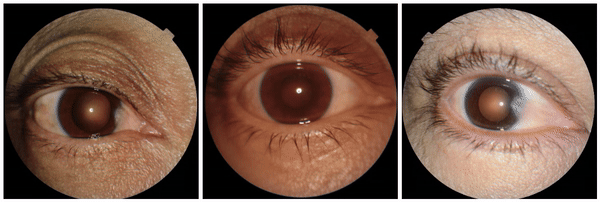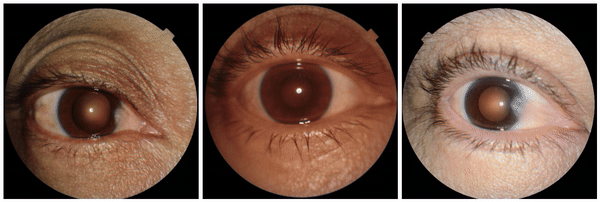
We also encountered a simultaneously surprising, retrospectively obvious, and prospectively thought-provoking result: increased eyeliner thickness and density correlated with lower hemoglobin levels. This finding likely reflects confounding factors within the dataset, as makeup usage is more common in females, who tend to have lower hemoglobin levels than males. This highlights the importance of considering dataset biases and quirks related to socio-cultural factors when interpreting ML model outputs.
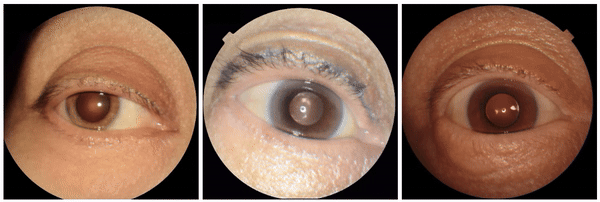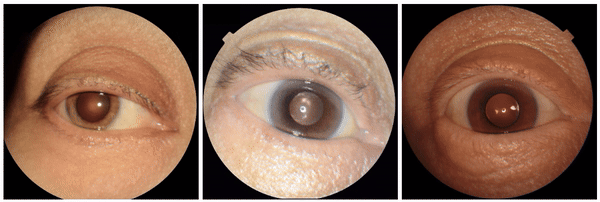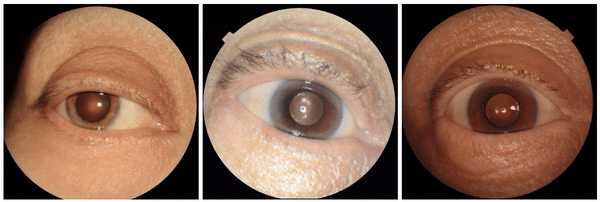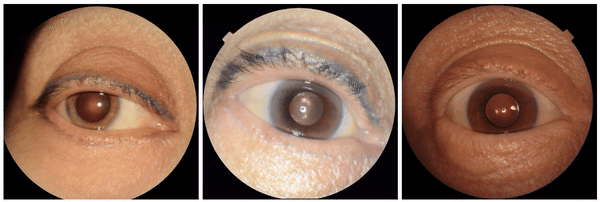
Examples of eyeliner, an attribute associated with lower predicted hemoglobin, a blood measurement lower in female individuals and the elderly.
Similarly, when examining the previously published research that AI models can identify race based on radiology images, one feature appeared related to the clavicle (collar bone) angle/position. This attribute is also seen when taking the CXR from different angles: posterior-anterior (PA, taken for outpatients standing up) versus anterior-posterior (AP, taken for patients on a hospital bed). In this dataset, we verified that the ratio of self-reported race differed for PA versus AP images with a directionality consistent with the observed attribute. This association may be due to a mix of factors including dataset gathering processes, healthcare access, or patient population aspects.
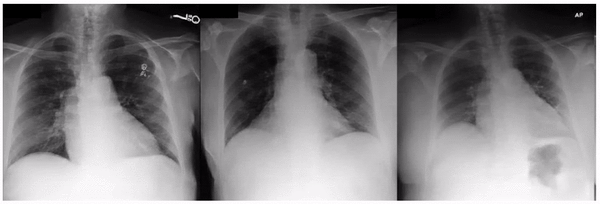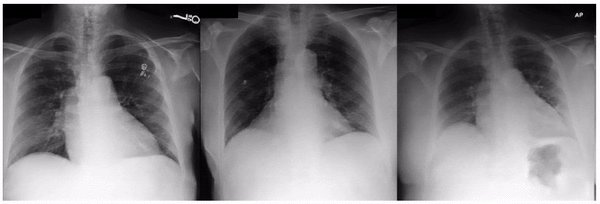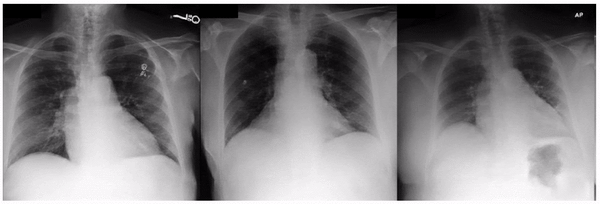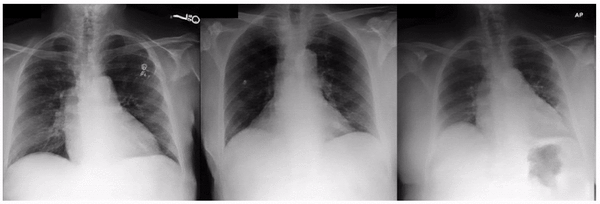
Examples of an apparent inferior displacement of the clavicles, associated with self-reported race. Animations are presented as a flickering between 2 frames to accentuate the difference.
Hypothesis generation, not causality
While our framework offers valuable interpretable insights, it's essential to acknowledge that it does not establish causality, but rather presents attributes for human interpretation and further investigation. Therefore, interdisciplinary collaboration is essential to ensure a rigorous interpretation. The expert panel review concluded that several observed associations might be influenced by unmeasured variables or complex relationships not captured by the model and unrelated to physiology, including the impact of structural and societal factors on health outcomes, dataset demographics or composition, and human interactions with technology. The expert panel also concluded that some associations might reveal novel phenomena and suggested research to support such discoveries.
We sum up these learnings in a general framework that we call Interdisciplinary Expert Panel to Advance Equitable Explainable AI. This framework can be used to guide interdisciplinary exploration of ML model results with the goal of helping to reduce bias, identify potential confounders, and identify opportunities for additional research where there are gaps in the literature on interpretation of ML model results. In turn, these insights can suggest opportunities for ML model improvement. For interpreting attributes, the associations were categorized as either a) known association; b) known in the clinical literature, could warrant further statistical examination; c) novel association, warrants further investigation; or d) strong likelihood of confounding or other bias present, further investigation highly recommended.
Conclusion
Our study demonstrates the potential of generative models to enhance the explainability of ML models in medical imaging. By combining technical advancements with interdisciplinary expertise, we can responsibly harness AI to uncover new knowledge, improve medical diagnostics, and address biases in healthcare. We encourage further research in this area, and emphasize the importance of collaboration between ML researchers, clinicians, and social scientists.
Acknowledgements
The authors thank Dr. Sreenivasa Raju Kalidindi and his team at Apollo Radiology International for their aid with the Apollo dataset, Andrew Sellergren and Zaid Nabulsi for help with CXR modeling infrastructure, Dr. Jorge Cuadros and Dr. Lauren P. Daskivich for their help with the EyePACS/LACDHS dataset, Elvia Figueroa and the LAC DHS TDRS program staff for data collection and program support, Nikhil Kookkiri and EyePACS staff for data collection and support, and Preeti Singh for support with dataset and annotation logistics. Finally, the authors would like to thank Avinash Varadarajan, Inbar Mosseri, and Yossi Gandelsman for early feedback on the project, Cameron Chen, Ivor Horn, and Lily Peng for providing feedback on the Lancet eBioMedicine manuscript, and Tiya Tiyasirichokchai for figure design. The authors would also like to thank Stephen Pfohl, Jessica Schrouff, Sami Lachgar, Lauren Winer, and Dale Webster for their feedback and support of the interdisciplinary expert panel framework manuscript.