How AI trained on birds is surfacing underwater mysteries
February 9, 2026
Lauren Harrell, Data Scientist, Google Research
We describe how Perch 2.0, Google DeepMind's bioacoustics foundation model, trained on birds and other terrestrial animal vocalizations, transfers ‘whale’ to underwater acoustics challenges with ‘killer’ performance.
Quick links
Underwater sound is critical for understanding the unseeable patterns of marine species and their environment. The ocean soundscape is full of mysterious noises and unfound discoveries. For example, the mysterious “biotwang” sound, recently attributed to the elusive Bryde’s whales by the U.S. National Oceanic and Atmospheric Administration (NOAA), illustrates the continuous challenge of new song types and species attributions being identified regularly.
Google has a long history of collaborating with external scientists on using bioacoustics for monitoring and protecting whales, including our original research models to detect humpback whale classifications and the release of our multi-species whale model in 2024. To keep up with this pace, Google’s approach to AI for bioacoustics is evolving to enable more efficient connections from new discoveries to scientific insights at scale. In August 2025, Google DeepMind released the latest Perch foundational bioacoustics model, Perch 2.0, a bioacoustics foundation model trained primarily on birds and other terrestrial vocalizing animals. Surprisingly, despite including no underwater audio in training, Perch 2.0 performed well as an embedding model for transfer learning in marine validation tasks.

A beluga whale — the “canary” of the sea. (Credit: Lauren Harrell)
In our latest paper, “Perch 2.0 transfers 'whale' to underwater tasks”, a collaboration between Google Research and Google DeepMind presented at the NeurIPS 2025 workshop on AI for Non-Human Animal Communications, we deep dive into these results. We show how this bioacoustics foundation model, trained mostly on bird data, can be used to enable and scale insights for underwater marine ecosystems, particularly for classifying whale vocalizations. We are also sharing an end-to-end tutorial in Google Colab for our agile modeling workflow, demonstrating how to use Perch 2.0 to create a custom classifier for whale vocalizations using the NOAA NCEI Passive Acoustic Data Archive through Google Cloud.
How bioacoustics classification works
If a pre-trained classification model, such as our multi-species whale model, already has the necessary labels and works well on a researcher’s dataset, it can be used directly to produce scores and labels for their audio data. However, to create a new custom classifier for newly discovered sounds or to improve accuracy on new data, we can leverage transfer learning instead of building a new model from scratch. This approach drastically reduces the amount of computation and experimentation needed to create a new custom classifier.
In bioacoustics transfer learning, the pre-trained model (such as Perch 2.0) is used to produce embeddings for each window of audio. These embeddings reduce the large audio data into a much smaller array of features that serve as input for a simple classifier. To create a new custom model for any set of labeled audio data, we apply the pre-trained model to the audio data to get the embeddings, which are used as the input features for a logistic regression classifier. Instead of learning all of the parameters for a deep neural network, we now only need to learn new parameters for the last step of logistic regression, which is much more efficient for both the researcher’s time and computational resources.
Evaluation
We evaluated Perch 2.0 using a few-shot linear probe on marine tasks, such as distinguishing different baleen whale species or different killer whale subpopulations. Its performance was compared against pre-trained models that are supported in our Perch Hoplite repository for agile modeling and transfer learning. They include Perch 2.0, Perch 1.0, SurfPerch, and the multispecies whale model.
For underwater data evaluation, we used three datasets: NOAA PIPAN, ReefSet, and DCLDE.
- NOAA PIPAN: An annotated subset of the NOAA NCEI Passive Acoustic Data Archive from the NOAA Pacific Islands Fisheries Science Center recordings. It includes labels used in our prior whale models as well as new annotations for baleen species such as common minke whale, humpback whale, sei whale, blue whale, fin whale, and Bryde’s whale.
- ReefSet: Developed for SurfPerch model training, this dataset leverages data annotations from the Google Arts and Culture project: Calling in Our Corals. It includes a mix of biological reef noises (croaks, crackles, growls), specific species/genera classes (e.g., damselfish, dolphins, and groupers), and anthropomorphic noise and wave classes.
- DCLDE: This dataset is evaluated using three different label sets:
- Species: For distinguishing between killer whales, humpbacks, abiotic sounds, and unknown underwater sounds (with some uncertainty in killer whale and humpbacks labels).
- Species Known Bio: For certain labels of killer whales and humpbacks.
- Ecotype: For distinguishing between killer whale subpopulations (ecotypes), including Transient/Biggs, Northern Residents, Southern Residents, Southeastern Alaska killer whales, and offshore killer whales.
In this protocol, for a given target dataset with labeled data, we compute embeddings from each of the candidate models. We then select a fixed number of examples per class (4, 8, 16, or 32), and train a simple multi-class logistic regression model on top of the embeddings. We use the resulting classifier to compute the area under the receiver-operating characteristic curve (AUC_ROC), where values closer to 1 indicate a stronger ability to distinguish between classes. This process simulates using a given pre-trained embedding model to create a custom classifier from a small number of labelled examples.
Our results show that more examples per class improve performance across all the models, except on ReefSet data, where performance is high even with only four examples per class for all models, except the multispecies whale model. Notably, Perch 2.0 is consistently either the top or second-best performing model for each dataset and sample size.
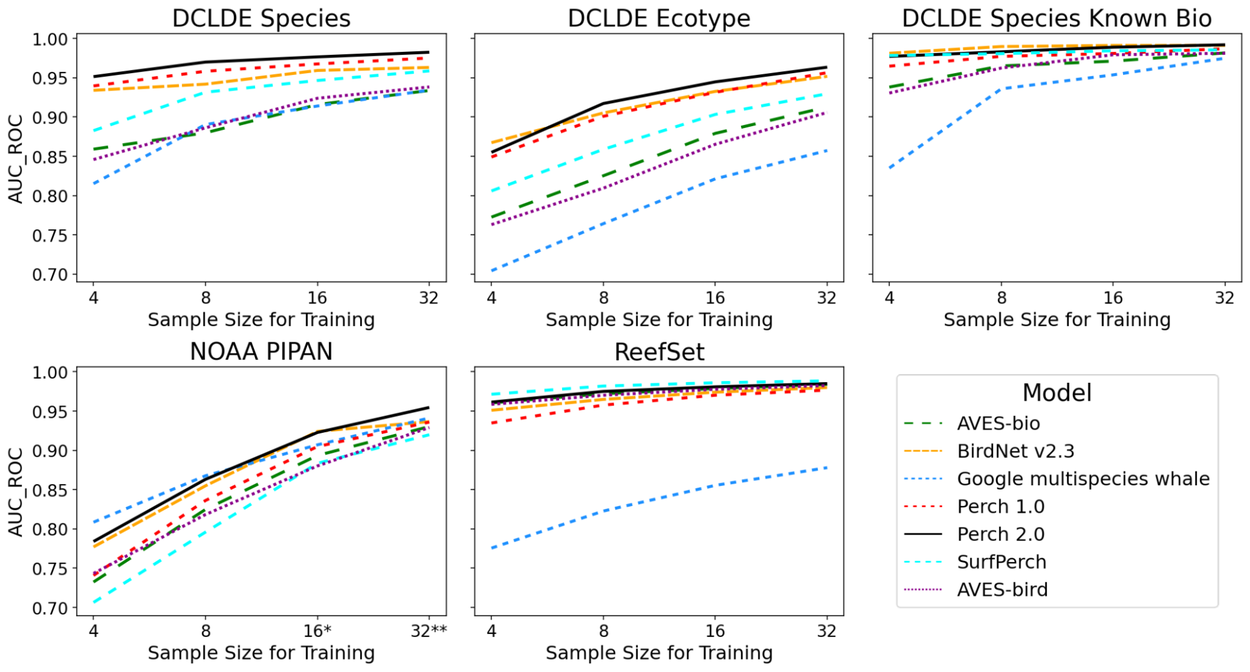
Performance of trained models on marine datasets, varying the number (k) of training examples per-class. Higher values of AUC_ROC indicate improved classification performance.*Class “Bm” dropped for k = 16; **Classes ‘Bm’ and ‘Be’ dropped for k = 32 in NOAA PIPAN data.
We also compare Perch 2.0 to embeddings from AVES-bird and AVES-bio (Earth Species Project transformer bioacoustics models trained on birds and biological sounds, respectively) and BirdNet v2.3 from K. Lisa Yang Center for Conservation Bioacoustics at the Cornell Lab of Ornithology. Perch 2.0 outperforms AVES-bird and AVES-bio on most underwater tasks, but there are other pre-trained models that also perform well that weren’t trained on underwater audio.
How does Perch 2.0 work so 'whale'?
We offer a few possible reasons for this transfer performance from our model trained primarily on birds to underwater sounds. First, prior research shows that larger models with extensive training data generalize better, allowing our bioacoustics model to perform well even on downstream tasks classifying sounds for species and sounds not included in the training dataset. Additionally, the challenge of classifying similar bird calls (the “bittern lesson”) forces the model to learn detailed acoustic features that can then be informative for other bioacoustics tasks. For example, there are 14 species of doves in North America, each with their own subtly distinct “coo” sound. A model that extracts the features that can distinguish between each species-specific “coo” is likely to isolate features that can help separate other sound classes. Finally, feature transfer across different types of species could also be related to the sound production mechanism itself, where a variety of species — including birds and marine mammals — have evolved similar means of sound production.
A high performing model will have embeddings that are informative and linearly separable for the applied target classes. To visualize, we plot a summary of embeddings from each model using a procedure called tSNE, where different colors represent different classes. A highly informative model will show distinct clusters for each class, whereas the classes will be more intermixed in a less informative model (such as the Google multi-species whale model). While almost all models show some distinct clusters of points for Southern Resident killer whales (KW_SRKW) and Southern Alaskan Residents (KW_SAR), the resulting embeddings of sounds from Northern Resident killer whales (KW_NRKW), Transient killer whales (KW_TKW), and Offshore killer whales (KW_OKW) are intermingled in models such as AVES-bio, AVES-bird, and SurfPerch, but are more clearly distinguished in BirdNet v2.3 and Perch 2.0.
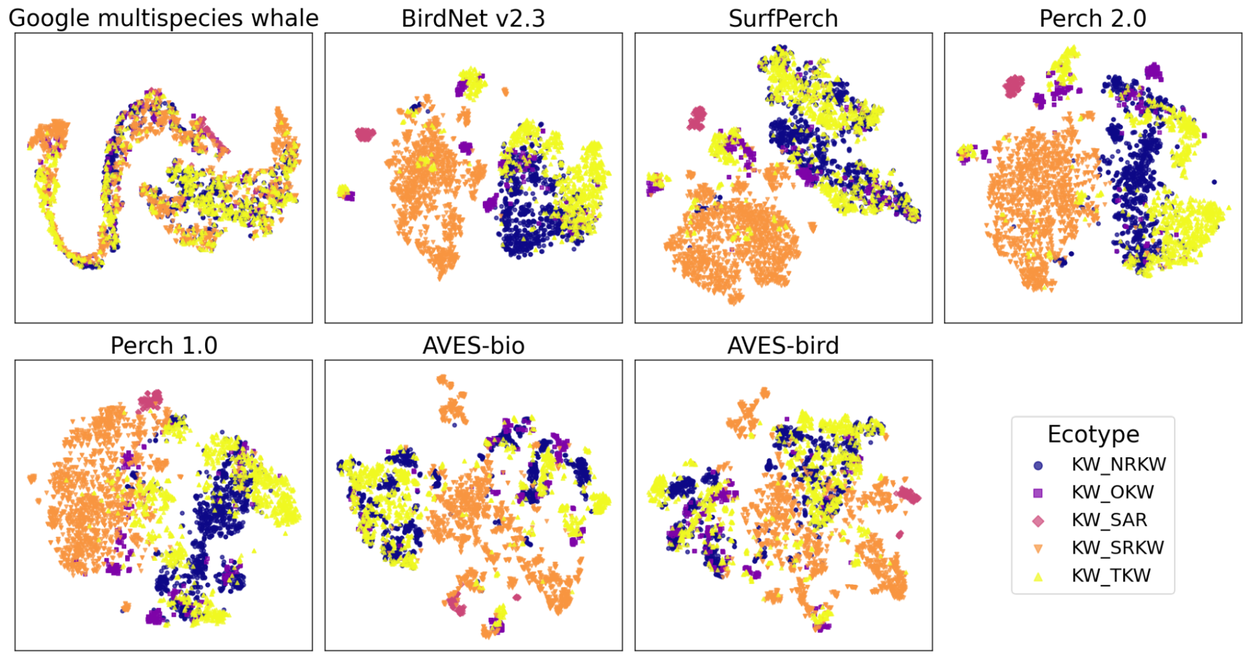
tSNE plots of the embeddings from each model on the DCLDE 2026 Ecotype dataset, which contains five ecotype variants of the killer whale (orca) species. Plots were generated with sci-kit learn PCA and tSNE libraries, with embeddings first projected to 32 dimension vectors prior to tSNE being applied.
Looking ahead
The Google DeepMind Perch team, in collaboration with Google Research and external partners, has pioneered an agile modeling approach for bioacoustics to create a custom classifier from a small number of labelled examples within a couple hours. In order to support both Google Research partners as well as the broader cetacean acoustics community, we have created an end-to-end demo for working with the NOAA data from the Passive Acoustic Archive dataset hosted on Google Cloud, updating our prior tutorials using the more efficient Perch Hoplite databases for managing embeddings.
Acknowledgements
The Perch Team, which developed the Perch 2.0 model and is part of Google DeepMind, includes Tom Denton, Bart van Merriënboer, Vincent Dumoulin, Jenny Hamer, Isabelle Simpson, Andrea Burns, and Lauren Harrell (Google Research). Special thanks to Ann Allen (NOAA Pacific Islands Fisheries Center) and Megan Wood (Saltwater Inc. in support of NOAA) for providing additional annotations used in the NOAA PIPAN dataset, Dan Morris (Google Research) and Matt Harvey (Google DeepMind).