Understanding Transfer Learning for Medical Imaging
December 6, 2019
Posted by Maithra Raghu and Chiyuan Zhang, Research Scientists, Google Research
Quick links
As deep neural networks are applied to an increasingly diverse set of domains, transfer learning has emerged as a highly popular technique in developing deep learning models. In transfer learning, the neural network is trained in two stages: 1) pretraining, where the network is generally trained on a large-scale benchmark dataset representing a wide diversity of labels/categories (e.g., ImageNet); and 2) fine-tuning, where the pretrained network is further trained on the specific target task of interest, which may have fewer labeled examples than the pretraining dataset. The pretraining step helps the network learn general features that can be reused on the target task.
This kind of two-stage paradigm has become extremely popular in many settings, and particularly so in medical imaging. In the context of transfer learning, standard architectures designed for ImageNet with corresponding pretrained weights are fine-tuned on medical tasks ranging from interpreting chest x-rays and identifying eye diseases, to early detection of Alzheimer’s disease. Despite its widespread use, however, the precise effects of transfer learning are not yet well understood. While recent work challenges many common assumptions, including the effects on performance improvement, contribution of the underlying architecture and impact of pretraining dataset type and size, these results are all in the natural image setting, and leave many questions open for specialized domains, such as medical images.
In our NeurIPS 2019 paper, “Transfusion: Understanding Transfer Learning for Medical Imaging,” we investigate these central questions for transfer learning in medical imaging tasks. Through both a detailed performance evaluation and analysis of neural network hidden representations, we uncover many surprising conclusions, such as the limited benefits of transfer learning for performance on the tested medical imaging tasks, a detailed characterization of how representations evolve through the training process across different models and hidden layers, and feature independent benefits of transfer learning for convergence speed.
Performance Evaluation
We first performed a thorough study on the effect of transfer learning on model performance. We compared models trained from random initialization and applied directly on tasks to those pretrained on ImageNet that leverage transfer learning for the same tasks. We looked at two large scale medical imaging tasks — diagnosing diabetic retinopathy from fundus photographs and identifying five different diseases from chest x-rays. We evaluated various neural network architectures including both standard architectures popularly used for medical imaging (ResNet50, Inception-v3) as well as a family of simple, lightweight convolutional neural networks that consist of four or five layers of the standard convolution-batchnorm-ReLU progression, or CBRs.
The results from evaluating all of these models on the different tasks with and without transfer learning give us four main takeaways:
- Surprisingly, transfer learning does not significantly affect performance on medical imaging tasks, with models trained from scratch performing nearly as well as standard ImageNet transferred models.
- On the medical imaging tasks, the much smaller CBR models perform at a level comparable to the standard ImageNet architectures.
- As the CBR models are much smaller and shallower than the standard ImageNet models, they perform much worse on ImageNet classification, highlighting that ImageNet performance is not indicative of performance on medical tasks.
- The two medical tasks are much smaller in size than ImageNet (~200k vs ~1.2m training images), but in the very small data regime, there may only be a few thousand training examples. We evaluated transfer learning in this very small data regime, finding that while there was a larger gap in performance between transfer and training from scratch for large models (ResNet) this was not true for smaller models (CBRs), suggesting that the large models designed for ImageNet might be too overparameterized for the very small data regime.
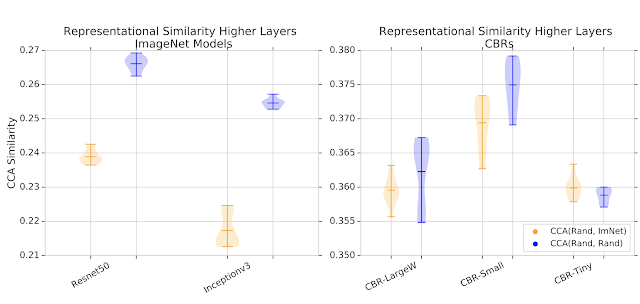
We next study the degree to which transfer learning affects the kinds of features and representations learned by the neural networks. Given the similar performance, does transfer learning result in different representations from random initialization? Is knowledge from the pretraining step reused, and if so, where? To find answers to these questions, this study analyzes and compares the hidden representations (i.e., representations learned in the latent layers of the network) in the different neural networks trained to solve these tasks. This quantitative analysis can be challenging, due to the complexity and lack of alignment in different hidden layers. But a recent method, singular vector canonical correlation analysis (SVCCA; code and tutorials), based on canonical correlation analysis (CCA), helps overcome these challenges, and can be used to calculate a similarity score between a pair of hidden representations.
Similarity scores are computed for some of the hidden representations from the top latent layers of the networks (closer to the output) between networks trained from random initialization and networks trained from pretrained ImageNet weights. As a baseline, we also compute similarity scores of representations learned from different random initializations. For large models, representations learned from random initialization are much more similar to each other than those learned from transfer learning. For smaller models, there is greater overlap between representation similarity scores.
When we combine the results of all the experiments from the paper, we can assemble a table summarizing how much representations change through training on the medical task across (i) transfer learning, (ii) model size and (iii) lower/higher layers.
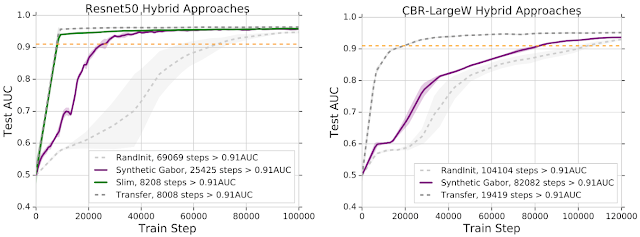
One consistent effect of transfer learning was a significant speedup in the time taken for the model to converge. But having seen the mixed results for feature reuse from our representational study, we looked into whether there were other properties of the pretrained weights that might contribute to this speedup. Surprisingly, we found a feature independent benefit of pretraining — the weight scaling.
We initialized the weights of the neural network as independent and identically distributed (iid), just like random initialization, but using the mean and variance of the pretrained weights. We called this initialization the Mean Var Init, which keeps the pretrained weight scaling but destroys all the features. This Mean Var Init offered significant speedups over random initialization across model architectures and tasks, suggesting that the pretraining process of transfer learning also helps with good weight conditioning.
 |
| Filter visualization of weights initialized according to pretrained ImageNet weights, Random Init, and Mean Var Init. Only the ImageNet Init filters have pretrained (Gabor-like) structure, as Rand Init and Mean Var weights are iid. |
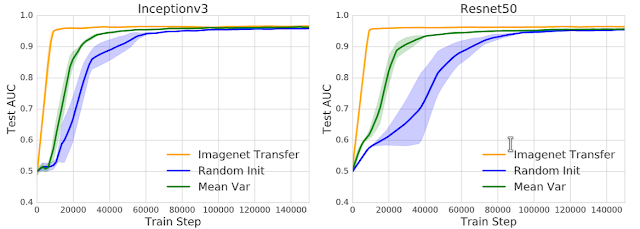 |
| Learning curves comparing the convergence speed with AUC on the test set. Using only the scaling of the pretrained weights (Mean Var Init) helps with convergence speed. The figures compare the standard transfer learning and the Mean Var initialization scheme to training from random initialization. |
Gabor filters (pictured below). Using just these (synthetic) weights offers significant speedups, and hints at many other creative hybrid approaches.
Transfer learning is a central technique for many domains. In this paper we provide insights on some of its fundamental properties in the medical imaging context, studying performance, feature reuse, the effect of different architectures, convergence and hybrid approaches. Many interesting open questions remain: How much of the original task has the model forgotten? Why do large models change less? Can we get further gains matching higher order moments of pretrained weight statistics? Are the results similar for other tasks, such as segmentation? We look forward to tackling these questions in future work!
Acknowledgements
Special thanks to Samy Bengio and Jon Kleinberg, who are co-authors on this work. Thanks also to Geoffrey Hinton for helpful feedback.
-
Labels:
- Machine Intelligence
Quick links
×
❮
❯