Tx-LLM: Supporting therapeutic development with large language models
October 9, 2024
Eric Wang, Student Researcher, Google DeepMind, and Dale Webster, Director, Google Research
Introducing Tx-LLM, a language model fine-tuned to predict properties of biological entities across the therapeutic development pipeline, from early-stage target discovery to late-stage clinical trial approval.
Quick links
Most candidates for therapeutic drugs fail clinical trials. Even if successful, they typically require 10–15 years and $1–2 billion to develop. A major reason for this is that the development pipeline contains many steps and many independent criteria that a therapeutic must satisfy. For example, a therapeutic should interact with its specific target but not other entities, producing the desired functional improvement but not off-target toxicity. Additionally, it should be able to travel to its desired destination, be cleared out of the body in an appropriate amount of time, and be suitable for manufacturing at scale. As one can imagine, measuring these properties experimentally is expensive and takes a long time, leaving an opportunity for an alternative approach: using machine learning (ML) to predict these properties quickly and efficiently.
To that end, we introduce Tx-LLM, a large language model (LLM) fine-tuned from PaLM-2, to predict properties of many entities (e.g., small molecules, proteins, nucleic acids, cell lines, diseases) that are relevant to therapeutic development. Tx-LLM is trained on 66 drug discovery datasets ranging from early-stage target gene identification to late-stage clinical trial approval, and therefore, is best suited to research on therapeutic applications. With a single set of weights, Tx-LLM achieved competitive performance with state-of-the-art models on 43 out of the 66 tasks and exceeded them on 22. Interestingly, we also observed that Tx-LLM exhibited abilities to combine molecular information with textual information as well as to transfer capabilities between tasks with diverse types of therapeutics. Overall, Tx-LLM is a single model that may be useful throughout the development of therapeutic drugs pipeline.
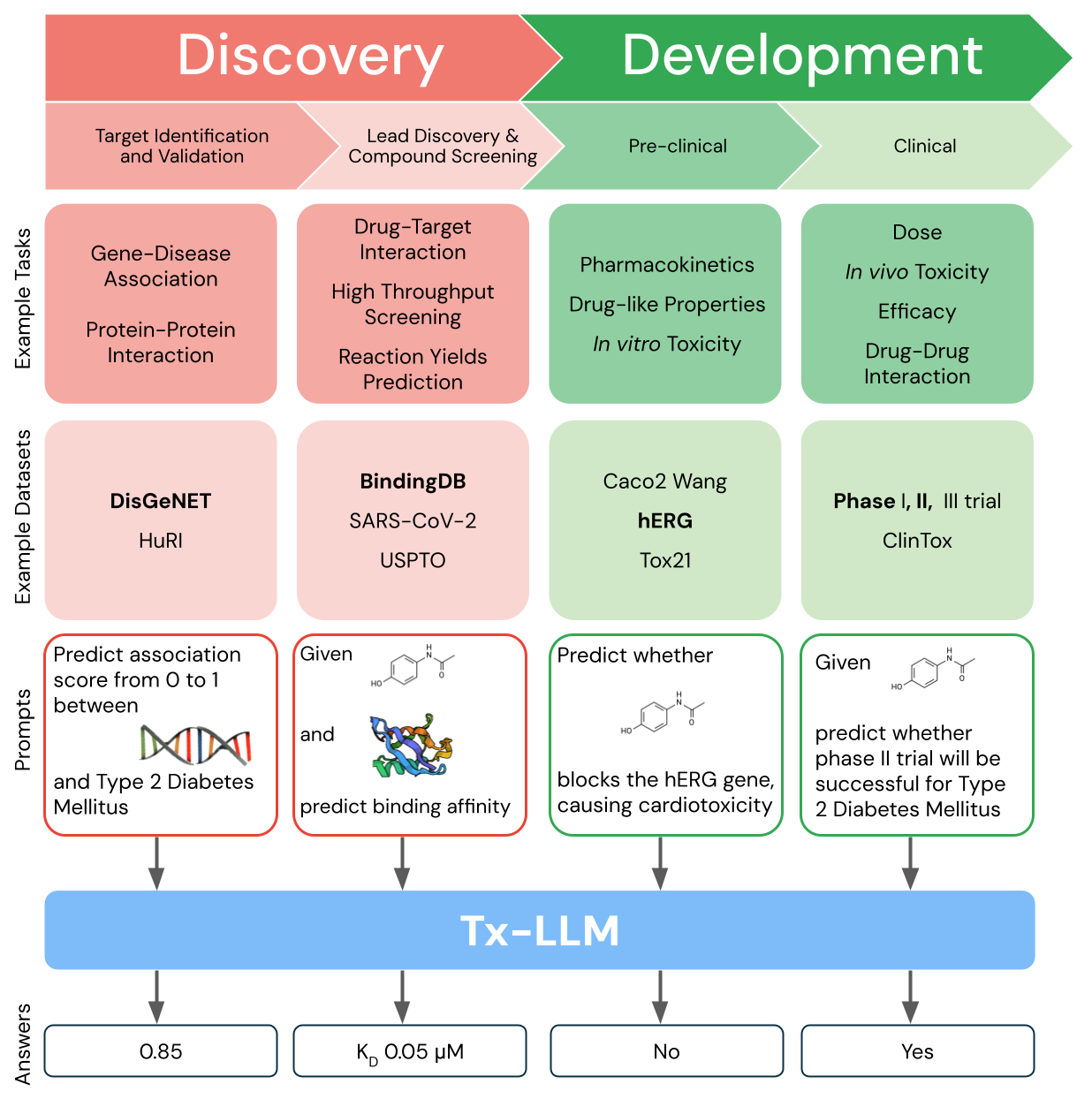
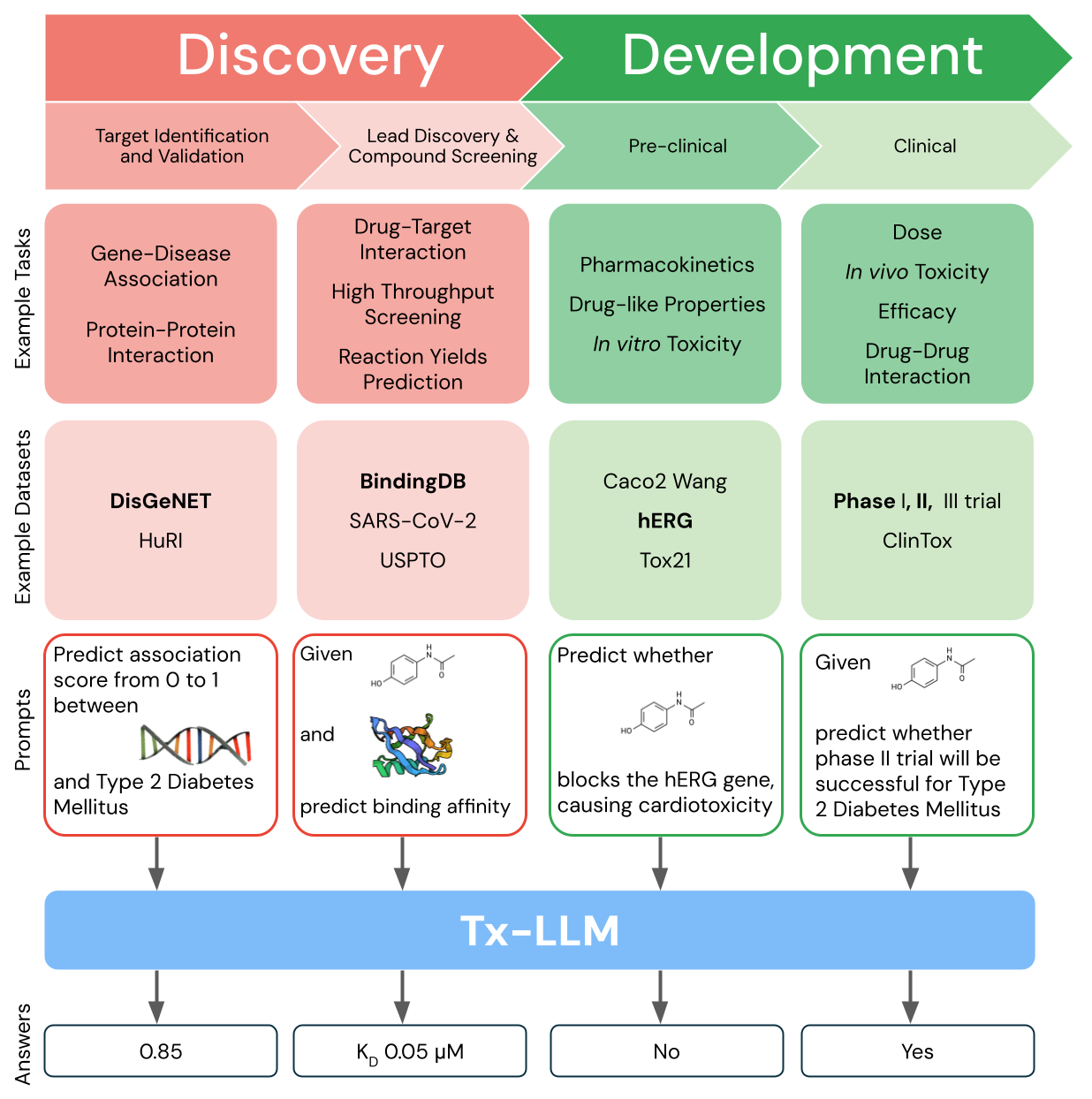
Tx-LLM is a single model that is fine-tuned to predict properties for tasks related to therapeutic development, ranging from early-stage target identification to late-stage clinical trial approval.
Curating the Therapeutics Instruction Tuning (TxT) collection for fine-tuning LLMs
Gathering data throughout the development pipeline is key to training the Tx-LLM model. We leveraged data from the Therapeutic Data Commons (TDC), a public collection of drug discovery datasets for training ML models, and processed 66 tasks most relevant to drug discovery into instruction-answer formats suitable for LLMs. Our collection, referred to as Therapeutics Instruction Tuning (TxT), structures each prompt with instructions, a context, a question, and an answer. Few-shot exemplars were also included in the question to enable in-context learning. TxT tasks can be broken down into three types:
- classification, which is framed as a multiple-choice question (e.g., output whether a drug is [A] non-toxic or [B] toxic)
- regression (e.g., output the binding affinity of a drug to a protein)
- generation (e.g., output which molecules were used in a chemical reaction)
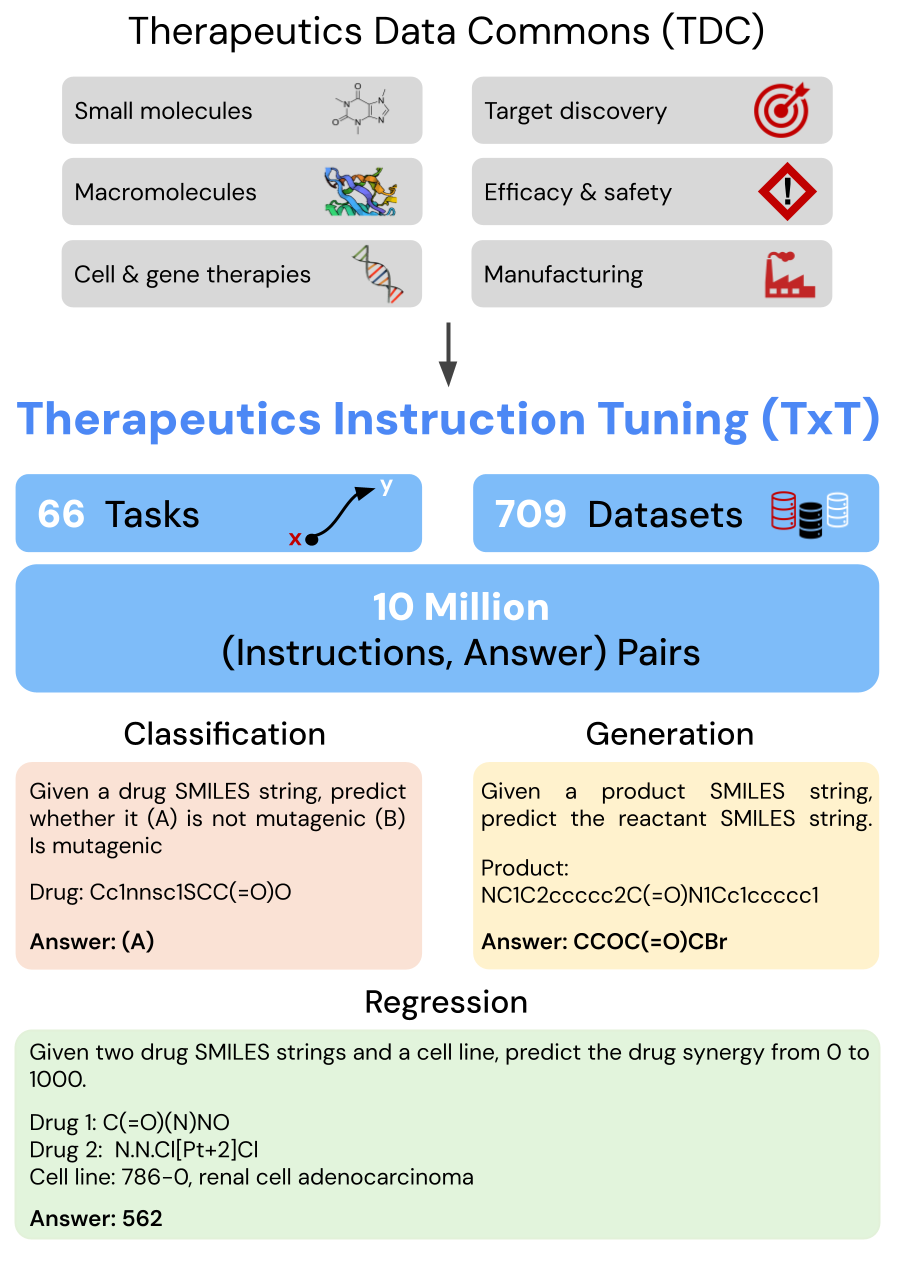
We curated Therapeutics Instruction Tuning (TxT), a collection of 709 datasets spanning 66 tasks related to therapeutic development, by reformatting tabular data from the Therapeutic Data Commons (TDC) into instruction-answer prompts for LLM training. The prompts shown here are shortened for brevity, and a full prompt is shown below (green).
TxT extends beyond the data found in TDC in a few notable ways. The context in each prompt supplies additional information that allows a generalist model to distinguish subtasks (e.g., training the model to predict toxicity in multiple different assays). Furthermore, some features in our dataset, such as cell lines, are represented directly as text as opposed to mathematical objects, such as gene expression vectors, where each entry of the vector is how much the cell expresses a particular gene. As we’ll see later, this representation also allows Tx-LLM to leverage its pre-training on natural language.
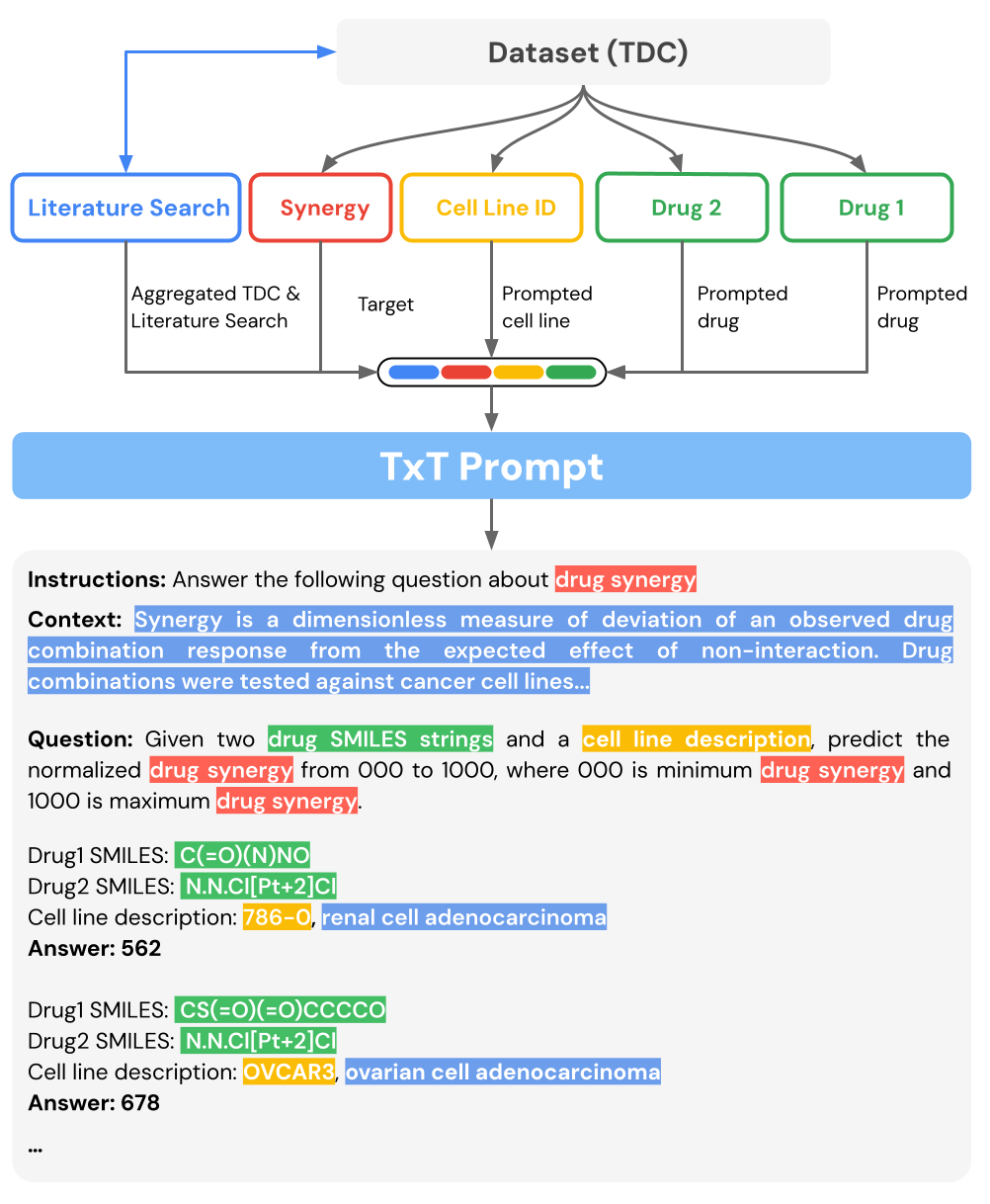
TxT uses data from TDC, supplemented with information from the literature, to construct a prompt.
Training Tx-LLM on TxT can outperform state-of-the-art specialist models
We then evaluated Tx-LLM on TDC datasets and compared it with existing state-of-the-art specialist models. Tx-LLM achieved competitive performance on 43 out of 66 tasks and exceeded their performance on 22. In many cases, we also observed that Tx-LLM was effective at predicting numeric values, which was somewhat surprising since LLMs have previously been observed to struggle with mathematical tasks. Binning the predictions into integers between 0 and 1000 may have helped with this, as it keeps the prediction format consistent and independent of units.
One area in which Tx-LLM was particularly effective was the combination of small molecules and text (e.g., given a drug and a disease name in a clinical trial, predict whether the drug would be approved). Tx-LLM actually outperformed current top models for the large majority of tasks involving these small molecules. This is likely because Tx-LLM was pre-trained on text including information about various diseases, so some context was already encoded in its weights.
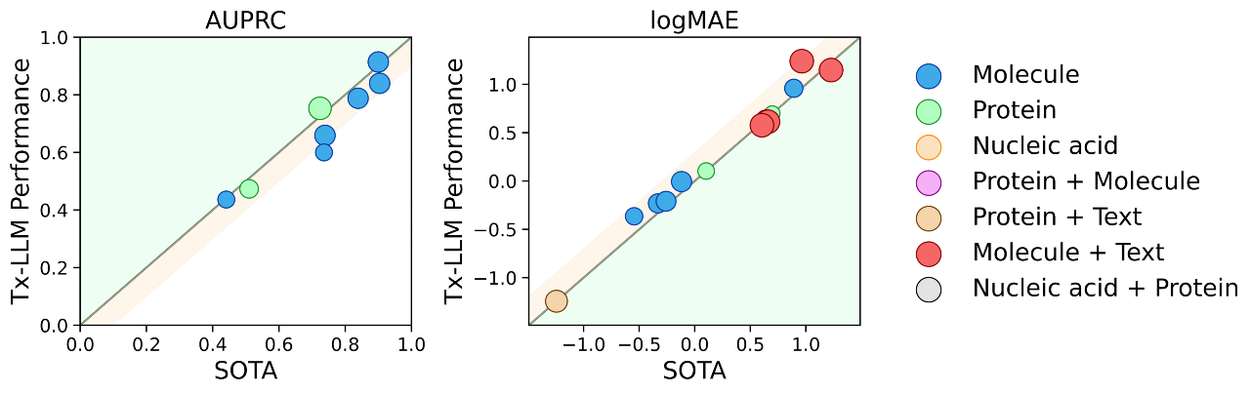
With a single set of weights, Tx-LLM is competitive with or exceeds state-of-the-art (SOTA) performance for binary classification and regression tasks encompassing diverse types of therapeutics (“Text” indicates cell lines or diseases represented using their names). Shaded green areas indicate stronger performance than existing state-of-the-art models, and shaded orange areas indicate near–state-of-the-art performance (within 10%). For results on all tasks, see the Tx-LLM paper.
Probing the successes of Tx-LLM
We then carried out an ablation study to determine what makes Tx-LLM work. Increasing the model size significantly improved performance and removing the prompt’s context significantly degraded performance, although changing the few-shot exemplars had no impact. Additionally, a contamination analysis with the PaLM-2 training data showed little overlap, and removing the overlapping examples had no impact on performance.
Interestingly, we observed positive transfer between tasks involving proteins and tasks involving small molecules. To study this, we compared Tx-LLM trained on all datasets with a version of Tx-LLM trained only on datasets involving small molecules, and we found that the version trained on all datasets performed better on tasks involving only small molecules, even though proteins and small molecules are quite different.
That said, Tx-LLM is still less effective than the top specialist models for many tasks, and experimental validation will remain a crucial part of the therapeutic development process even as ML models continue to evolve. Tx-LLM is not yet instruction-tuned to follow natural language, so it cannot explain its predictions to the user. Developing this functionality, as well as integrating the Gemini family of models, remains an exciting area to improve Tx-LLM.
Conclusions
Our overarching goal is to expedite the therapeutic development process, which is currently challenged by decade-long timelines and high cost. Tx-LLM represents a significant step forward — a single LLM fine-tuned on the Therapeutics Instruction Tuning, a comprehensive dataset spanning 66 tasks crucial to therapeutic development from beginning to end. Tx-LLM demonstrably achieves competitive or improved performance compared to existing specialist models, especially those combining molecular and textual information. As we look toward further improvements, we are excited by a future where Tx-LLM and similar models can streamline therapeutic development and ultimately improve human health.
Our team is assessing how we can make Tx-LLM’s capabilities available to external researchers. If you have a use case you’d like to explore or you’d just like to be kept informed of our progress, we’d love to hear from you! Understanding your use cases and interest informs our continued research.
Acknowledgements
Key contributors to this project include Eric Wang, Juan Manuel Zambrano Chaves, Shekoofeh Azizi, Vivek Natarajan, Tao Tu, Eeshit Dhavel Vaishnav, Byron Lee, S. Sara Mahdavi, Christopher Semturs, and David Fleet. We also thank David Belanger, Rory Pilgrim, Fan Zhang, Andrew Sellergren, Sami Lachgar, Lauren Winer, Maggie Shiels, Jessica Valdez, Jane Park, Jon Small, Aaron Abood, Rishad Patel, Uchechi Okereke, Annisah Um’rani, Alan Karthikesalingam, Anil Palepu, Juraj Gottweisto, Yun Liu, Dale Webster, Zoubin Ghahramani, Raia Hadsell, Joelle Barral, Pushmeet Kohli, Jon Shlens and Greg Corrado for their feedback and support throughout this project.





