Taking medical imaging embeddings 3D
October 21, 2024
Atilla Kiraly and Madeleine Traverse, Software Engineers, Google Research
Announcing the release of a new medical foundation tool for 3D CT volumes: CT Foundation. CT Foundation builds on our work in chest radiographs, dermatology and digital pathology, extending it to the realm of 3D volumes.
Over recent years, developers and researchers have made progress in efficiently building AI applications. Google Research has contributed to this effort by providing easy-to-use embedding APIs for radiology, digital pathology and dermatology to help AI developers train models in these domains with less data and compute. However, these applications have been restricted to 2D imaging, while physicians often use 3D imaging for complex diagnostic decision-making. For example, computed tomography (CT) scans are the most common 3D medical imaging modality, with over 70 million CT exams conducted each year in the USA alone. CT scans are often essential for a variety of critical patient imaging evaluations, such as lung cancer screening, evaluation for acute neurological conditions, cardiac and trauma imaging, and follow-up on abnormal X-ray findings. Because they are volumetric, CT scans are more involved and time-consuming for radiologists to interpret compared to 2D X-rays. Similarly, given their size and structure, CT scans also require more storage and compute resources for AI model development.
CT scans are commonly stored as a series of 2D images in the standard DICOM format for medical images. These images are then recomposed into a 3D volume for either viewing or further processing. In 2018, we developed a state-of-the-art chest lung cancer detection research model trained on low dose chest CT images. We’ve subsequently improved the model, tested it in clinically realistic workflows and extended this model to classify incidental pulmonary nodules. We’ve partnered with both Aidence in Europe and Apollo Radiology International in India to productionize and deploy this model. Building on this work, our team explored multimodal interpretation of head CT scans through automated report generation, which we described in our Med-Gemini publication earlier this year.
Based on our direct experience with the difficulties of training AI models for 3D medical modalities, coupled with CT’s importance in diagnostic medicine, we designed a tool that allows researchers and developers to more easily build models for CT studies across different body parts. Today we announce the release of CT Foundation, a new research medical imaging embedding tool that accepts a CT volume as input and returns a small, information-rich numerical embedding that can be used for rapidly training models with little data. We developed this model for research purposes only and as such it may not be used in patient care, and is not intended to be used to diagnose, cure, mitigate, treat, or prevent a disease. For example, the model and any embeddings may not be used as a medical device. Interested developers and researchers can request access to the CT Foundation API, and use it for research purposes at no cost. We have included a demo notebook on training a model for lung cancer detection using the publicly available NLST data from The Cancer Imaging Archive.
How CT Foundation works
CT Foundation handles all of the processing of a given CT volume in DICOM format and produces an information-rich embedding vector, a set of 1,408 numbers that summarize important information from the given CT volume. These embedding vectors include information about organs, tissues, and the presence of abnormalities. The CT Foundation API processes the raw DICOM images by sorting the individual slices, composing them into a volume, running model inference and returning CT embeddings. This saves users from needing to preprocess DICOM images.
These embeddings can then be used as an input for downstream classifier models, such as logistic regression or multilayer perceptrons. Using the embeddings, one can produce performant models on classification tasks with less data compared to initializing training from a generic 3D model. Because the end user only needs to train the final classifier, which is relatively small in size, they also save considerably on compute compared to fine-tuning a generic 3D model.
CT Foundation was developed using VideoCoCa, a video-text model designed for efficient transfer learning from 2D Contrastive Captioners (CoCa). CoCa models take text and images as input and encode them into a shared, language-aligned embedding space. They include a multimodal text decoder that can decode these embeddings into text tokens. CoCa models are trained to minimize two types of loss. The first is captioning loss, the loss between the original ground-truth captions of the training images and the ones decoded by the CoCa model. This focuses on the accuracy of the provided caption. The second is contrastive loss, which aims to minimize the distance between CoCa’s encodings of image-text pairs, resulting in a richer semantic understanding of the images. VideoCoCa extends an existing CoCa model by pooling together multiple frames to produce a compact representation of the entire set of sequence images.
CT Foundation was trained using over a half-million de-identified CT volumes that include a range of body parts from the head to extremities, each paired with their corresponding radiology reports. We first trained a medical image–specific 2D CoCa model and applied it as a basis for VideoCoCa. We then trained VideoCoCa with axial CT slices (sequence of CT slices that comprise the volume) coupled with radiology reports.
Left-sided nephrolithiasis and urolithiasis with stones in the left kidney, ureter, and ureterovesical junction as identified by a model created using CT Foundation.
Evaluating CT Foundation
To test CT Foundation’s utility and generalizability, we evaluated its data efficiency across seven classification tasks. This included six clinically relevant classification tasks spanning head, chest, and abdominopelvic regions, each involving the detection of abnormalities. These tasks were related to classifying: intracranial hemorrhage, calcifications in the chest and heart, lung cancer prediction in the chest, suspicious abdominal lesions, nephrolithiasis, and abdominal aortic aneurysm in abdominopelvic CTs. All binary labels with the exception of the lung cancer and hemorrhage tasks were automatically extracted from the clinical radiology reports, which were written by board certified radiologists. The lung cancer prediction task was drawn from NLST and used pathology-confirmed cancer outcomes within 2 years of the lung screening task for cancer positive labels. The hemorrhage task was labeled by board certified radiologists. We wanted to measure the model’s data-efficient classification performance. So we trained multi-layer perceptron models for increasing amounts of training data. The evaluation set to measure the performance across these perceptrons was held constant. We evaluated the six clinical tasks using the area under the receiver-operating characteristic curve (AUC) metric. AUC is measured between 0.0–1.0, where 1.0 is a perfect model and 0.5 represents random chance.
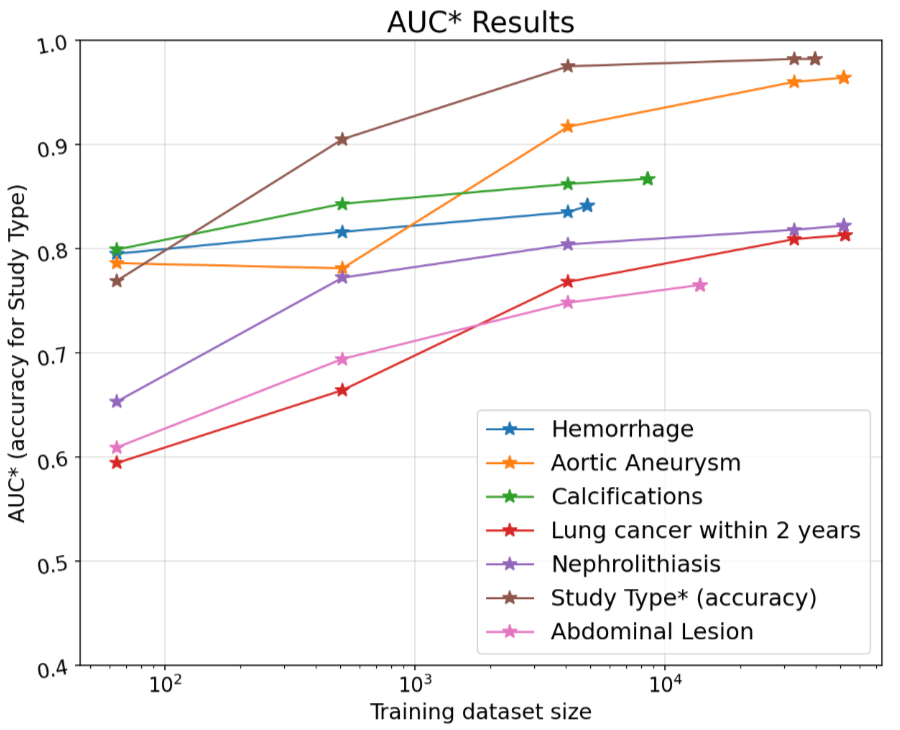
Performance results of non-linear models trained on CT Foundation embeddings across various CT studies and labels along with CT Study type classification results. Each model was trained only using the CPU with different sizes of training data and the evaluation subset was held constant. N refers to the total size of the evaluation set used to measure performance. Throughout each training run the positive ratio in the training set approximately matches the evaluation set. The lung cancer training and eval splits match those used for training and tuning by our previous publication.
Finally, as a more general task to illustrate CT Foundation’s utility for workflow applications, we included a higher-level body part classification task, which aimed to identify the anatomical region that was the target of the CT scan. The performance metric for this task was accuracy across eight different study types: Head/Neck, Neck, Spine, Heart, Angio, Chest, Abdomen and Pelvis, Extremities.
Given the relatively small size of the embeddings, each embedding is a 1,408 vector, only a CPU was needed to train the model, all within a Colab Python notebook. Even with limited training data, the models were able to reach over 0.8 AUC for all but one of the more difficult tasks.
Conclusion
With the release of our CT Foundation tool, we are also sharing a Python notebook to work with CT volumes and train and evaluate models. With its data-efficient and low-compute design, CT Foundation can enable rapid prototyping and research even with limited resources. The automated handling of DICOM format data eases CT modeling for both new and experienced researchers and developers. We're eager to see how researchers and developers utilize this tool and keen to receive community feedback on CT Foundation’s performance and use cases. Sign up here to get started!
Acknowledgements
We thank Google Health team members who led this research and made the public release possible, including Andrew Sellergren, Akshay Goel, Fereshteh Mahvar, Rory Pilgrim, Arnav Agharwal, Charles Lau, Yun Liu, Bram Sterling, Kenneth Philbrick, Lin Yang, Howard Hu, Nick George, Kristina Kozic, Gabby Espinosa, Fayaz Jamil, Aurora Cheung, Shruthi Prabhakara, Can Kirmizi, Daniel Golden, Shekoofeh Azizi, Christopher Kelly, Lauren Winer, Jenn Sturgeon, and Shravya Shetty. Significant contributions and input were also made by Dr. Greg Sorensen and his colleagues at DeepHealth and Dr. Raju and his colleagues at Apollo Radiology. We also thank the NIH Cancer Institute for making the LIDC and NLST data publicly available.



