Predicting fetal well-being from cardiotocography signals using AI
September 27, 2024
Mercy Asiedu, Research Scientist, Google Research, and Nichole Young-Lin, Clinical Lead, Google Health
We present our work on developing and evaluating a machine learning model for cardiotocography, to predict fetal well-being, and to understand what factors influence model performance.
Quick links
Cardiotocography (CTG) is a doppler ultrasound–based technique used during pregnancy and labor to monitor fetal well-being by recording fetal heart rate (FHR) and uterine contractions (UC). CTG can be done continuously or intermittently, with leads placed either externally or internally. External CTG involves the use of two sensors placed on the birthing parent’s belly: an ultrasound transducer placed above the fetal heart position to monitor FHR, and a tocodynamometer (pressure sensor) placed on the fundus of the uterus to measure UC.
Currently, providers interpret CTG recordings using guidelines like those from the National Institute of Child Health and Human Development (NICHD; guidelines) or the International Federation of Gynecologists and Obstetricians (FIGO; guidelines). These standards define different patterns in the CTG and FHR traces that may indicate fetal distress.
Today we present work from our recent paper, ”Development and evaluation of deep learning models for cardiotocography interpretation”, in which we describe research on our new machine learning (ML) model that will provide objective interpretation assistance to health providers to reduce burden and potentially improve fetal outcomes. Using an open-source CTG dataset, we develop end-to-end neural network-based models to predict measures of fetal well-being, including both objective (fetal arterial cord blood pH, i.e., fetal acidosis) and subjective (fetal Apgar scores) measures. Given the potential high stakes nature of the use-case if utilized in a clinical setting, we perform extensive evaluations to examine how the model performs with varying inputs, including FHR only, FHR+UC, and FHR+UC+Metadata.
This work builds a model development and evaluation pipeline to enable fetal well-being prediction that takes into account limited data, clinical metadata, and intermittent methods used in low-resource settings.
Improving CTG interpretation with deep neural networks
Presently, CTG and ultrasound are the primary means of evaluating fetal well-being in utero. Although CTG is routinely used in medical practice, its application to continuous intrapartum fetal monitoring is associated with a high false-positive rate with limited demonstration of improvements in fetal outcomes. This high false-positive rate has led to an increase in cesarean section and operative vaginal delivery rates with limited improvements in neonatal outcomes. This is likely due to the complexity of reading and interpreting the fetal heart tracings, and the subjective nature of visual interpretation methods and intra- and inter-observer variability when reading the tracings. These issues are exacerbated in low-resource facilities where access to skilled interpreters is even more limited.
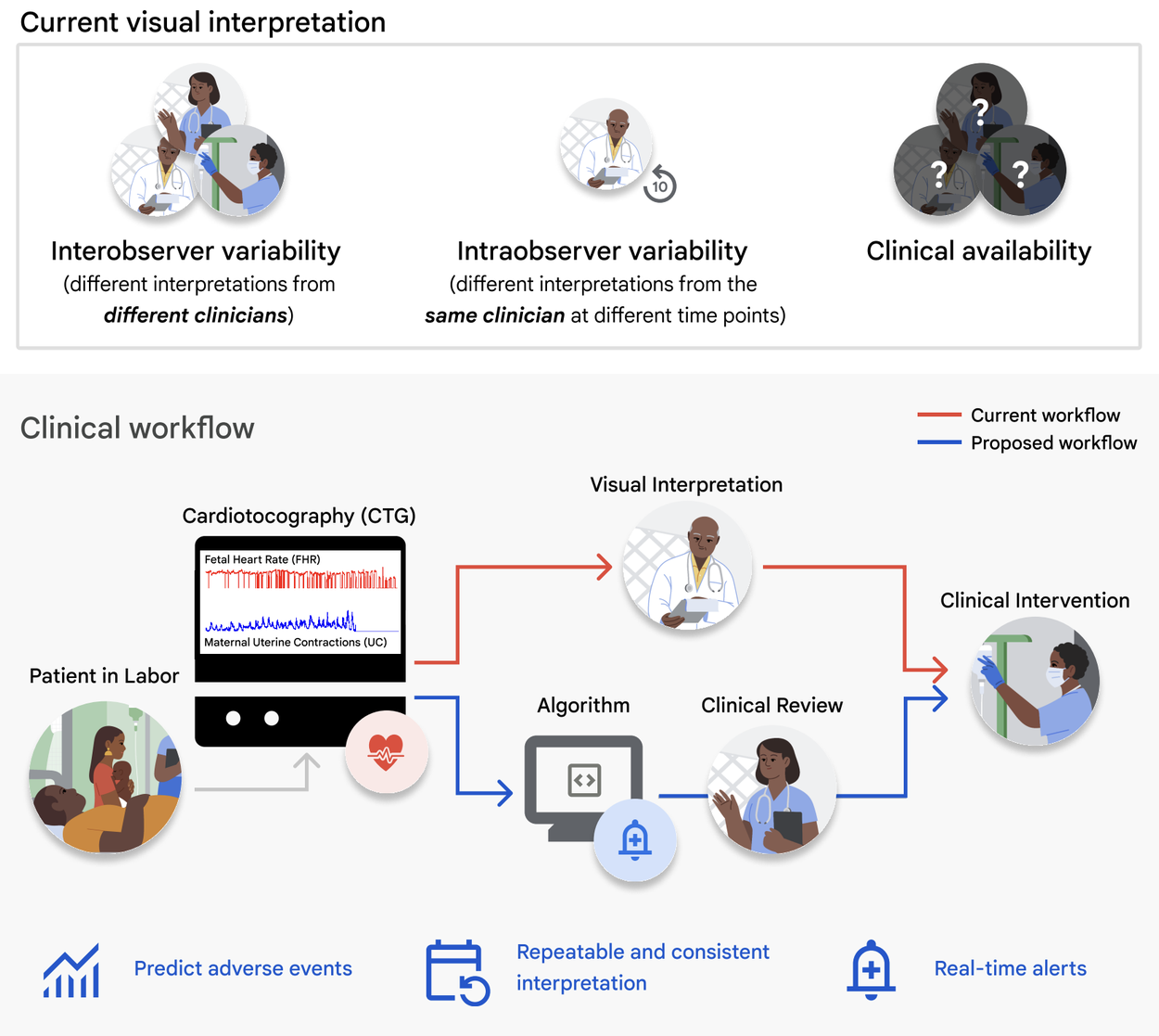
Top: Challenges of current visual CTG interpretation. Bottom: Proposed clinical use-case of deep learning algorithms for CTG interpretation and assistive clinical decision-making.
Current research methods using ML algorithms to classify abnormal CTGs typically use tabulated rules-based extraction of diagnostic features, such as summary statistics of fetal heart rate. While this approach has shown promise for improving clinical decision support, feature extraction reduces rich CTG information from the time series data. So, for CTG interpretation, there is a recent shift in focus to deep learning methods that use physiological time series data as input [1, 2, 3]. However these methods do not typically compare performance differences between objective and subjective ground truth labels, and do not explore the effects of intermittent measurements or clinical metadata.
Using an open license dataset, CTU-UHB Intrapartum Cardiotocography Database, which has 552 FHR and UC CTG signal pairs up to 90 minutes before delivery for a total of ~50,000 minutes of recordings:
- We highlight the feasibility of using deep learning methods to predict fetal hypoxia from CTG.
- We conduct evaluation studies to analyze the effect of:
- The choice of objective (arterial umbilical cord blood pH) vs. subjective (Apgar score) labels,
- The signal time interval used for training and testing,
- The evaluation of simulated low-resource environment intermittent signals on predictive performance.
Building on the current state-of the art models for CTG interpretation
Model architecture
We begin with the CTG-net network architecture, which convolves the paired FHR and UC input signals temporally before conducting a depthwise convolution to learn the relationship between them. We add the following methodological configurations:
- Architecture and hyperparameter optimization: We run a randomized convolution parameter and hyperparameter search to select the optimal model configurations and hyperparameters for the tasks.
- Single input variation: We develop a 1D convolutional neural network variation of the model that takes in only one signal to compare performance with either FHR or UC individually and explore the use of the individual signals.
- Addition of metadata: We add the clinical metadata as a vector to the input.
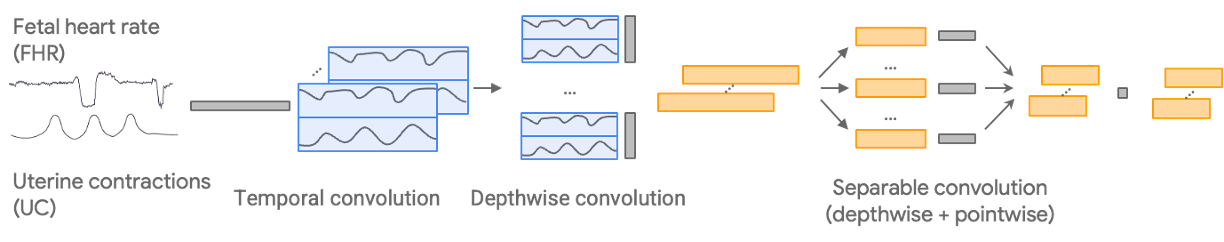
Development pipeline for the model, which uses FHR and UC inputs and generates a predicted outcome depending on the classification task.
Pre-processing
We create a pre-processing pipeline for input signals to improve data quality, smooth the signal, and account for gaps. This includes inputting missing measurements, random cropping (for pre-training and specific training evaluations), and additive multiscale noise for data augmentation and downsampling. This generates 4.3M minutes (n=496 patients) of signals for pre-training, ~150k minutes (n=496 patients) for training, and ~1,700 minutes (n=56 patients) for testing.
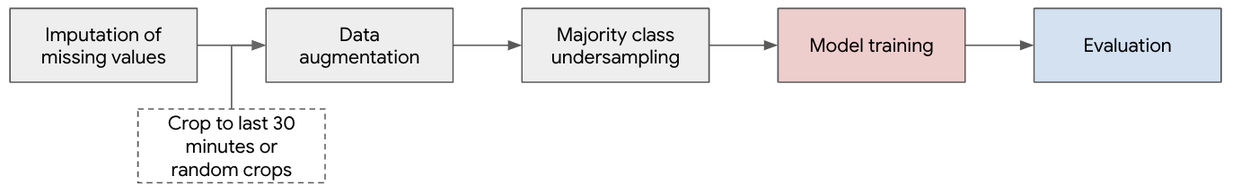
Pre-processing pipeline.
Pre-training
Given the limited number of patients in the open license dataset (n=552), we pre-train the model on cropped signal segments before the last 30 minutes and then fine-tune on the last 30 minutes of the test set, which we use as our primary time point of interest.
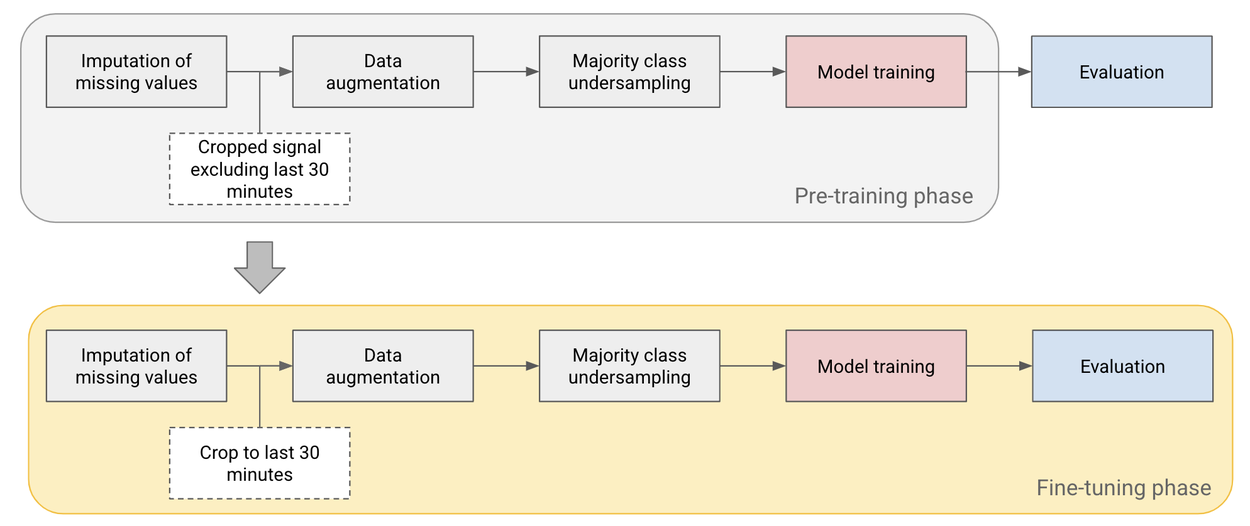
Pre-training and fine-tuning pipeline used to improve performance on small datasets.
Intermittent versus continuous CTG use cases
CTG use comes in two primary formats, intermittent and continuous. In most high-resource settings, continuous CTGs are used in the clinic throughout labor to continuously monitor fetal heart rate. These typically digital signals record uterine contractions and the fetal heart rate. However in low-resource settings, intermittent CTGs are often used, which may cover only about 30 minutes at any point during labor, and are then printed out for interpretation by the provider.
The open source data from CTU-UHB database came from a continuous CTG setting, in contrast with intermittent analog CTGs typically seen in low-resource settings. One of our key contributions is to understand how training and evaluating on intermittent time points impacts the model performance. We simulated intermittent settings as part of our evaluation process by splitting the 90 minute signals in the dataset into 30 minute signals and training and evaluating the model at different time points.
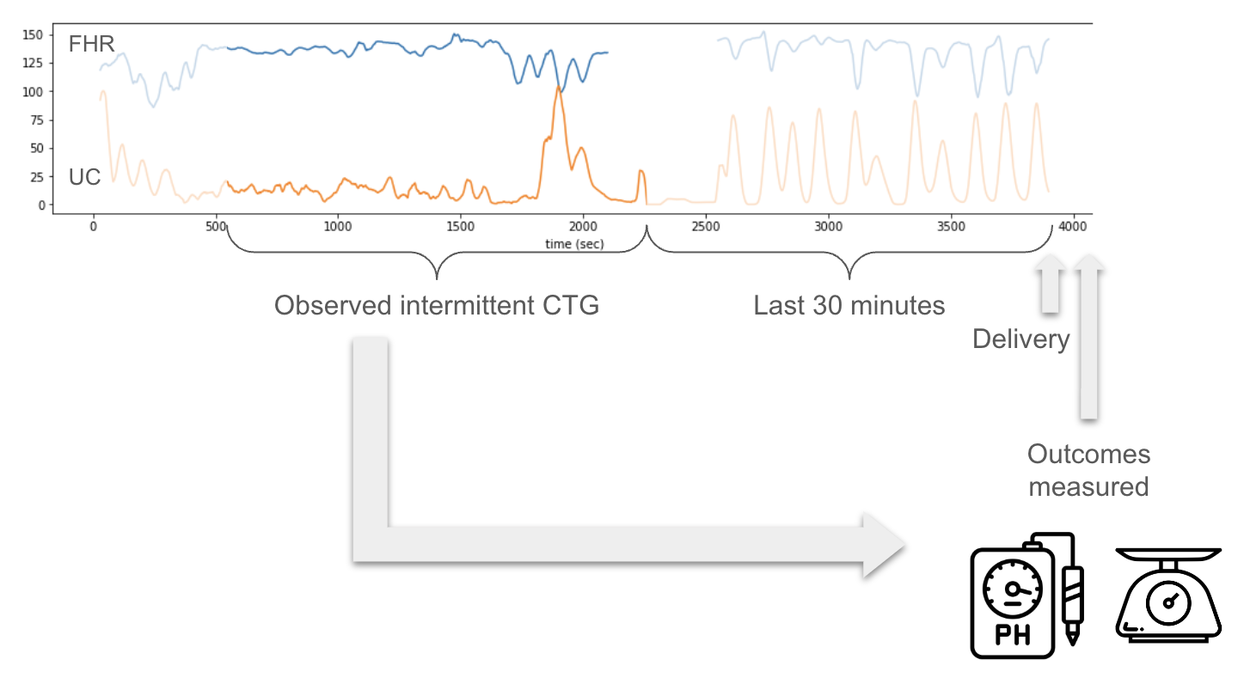
Intermittent CTG measurements are more likely to be the case in low-resource settings. Intermittent CTG is a 20–30 minute CTG measurement during labor at an unknown time point before birth. We simulate this setting in our evaluation to understand how models either trained or tested on intermittent datasets perform.
Predicting objective and subjective ground truth labels
Another key methodological contribution is our use of three outcome labels from the dataset:
- The arterial umbilical cord blood pH is an objective measurement, typically available in high-resource settings, that tracks fetal acidosis, which is an indication of fetal distress. The pH was considered abnormal if the score was less than 7.2.
- The Apgar score is a subjective measure (ranging from 0–10) recorded by a clinician after delivery that reflects the general health of the newborn. Apgar scores are the primary delivery outcome descriptor in low-resource settings due to their simplicity, cost-effectiveness, and the potential financial burden of umbilical cord blood pH analysis. The 1-minute Apgar score was considered abnormal if the score was less than 7.
- A label of “abnormal” if either Apgar or pH results were abnormal.
Evaluating model prediction robustness
For evaluation purposes we perform the following comparisons.
- Our performance on the dataset versus the state-of-the-art CTG-net model
- Apgar versus pH classification tasks
- FHR-only versus FHR+UC
- Base model using last 30 minutes of labor (continuous case) versus intermittent measurements
- Base model of FHR+UC versus FHR+UC+Metadata
- Subgroup performance of the base model (FHR+UC) with subgroups determined by binarizing clinical metadata (e.g., low/high maternal age, low/high gestational weeks at birth, frequent/infrequent signal gaps)
We find that our approach performs comparably to the reported AUROC in CTG-net, even though it is trained on a smaller dataset. When we train and evaluate both methods on the same dataset we find that our method improves model performance by 10+ AUROC percentage points.
| AUROC | ||
|
CTG-net* |
0.68 ± 0.03 |
– |
|
CTG-net (on the same dataset we used) |
0.57 ± 0.08 |
– |
|
Our Model† |
0.68 ± 0.07 |
0.27 (0.18) |
|
– |
0.45 (95% CI: 0.23-0.68) |
This table compares performance for the models, and clinicians on CTG data for binary classification tasks of abnormal fetal status (pH < 7.2 or Apgar at 1-min < 7). *=Mean and std reported over 10 random training seeds, †=Mean and std reported over 1,000 bootstrap samples
We find that combining FHR+UC achieves the highest model performance for both pH and Apgar classification. For the intermittent training and evaluation tasks, we find that the Apgar prediction task exhibits less robustness and more variability across different trained time periods, whereas the objective pH value is more stable. We also find that the pre-training step enables the highest model performance. Adding clinical metadata, such as maternal age and health status (e.g., pre-eclampsia / gestational diabetes), slightly improves model performance for pH, but less so for Apgar.
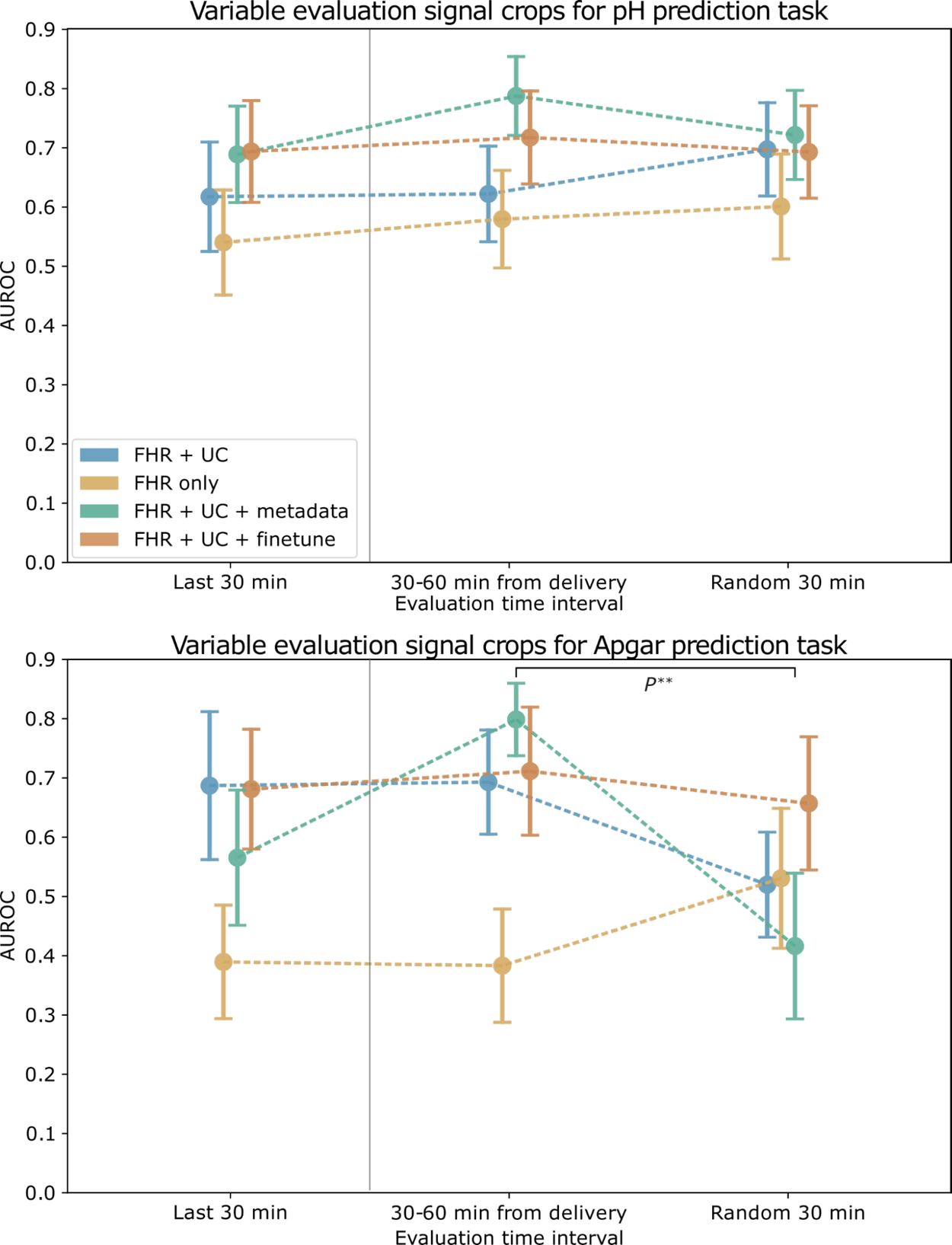
AUROC for pH (top) and Apgar (bottom) classification models evaluated on a cropped signal from different time intervals. Error bars depict the standard error, computed over 1,000 bootstrap samples. Markers to the left of the vertical gray line indicate the paradigm used to train and evaluate the baseline models.
Subgroup evaluations
We found significant differences in baseline performance between subgroups with frequent and infrequent UC signals gaps (UC missing) for pH prediction and for subgroups with frequent and infrequent FHR signal gaps (FHR missing) for Apgar prediction. With metadata, the performance disparities observed with pH prediction were mitigated. However, including metadata increased the AUROC performance disparities for demographic and clinical-related subgroups on this task.
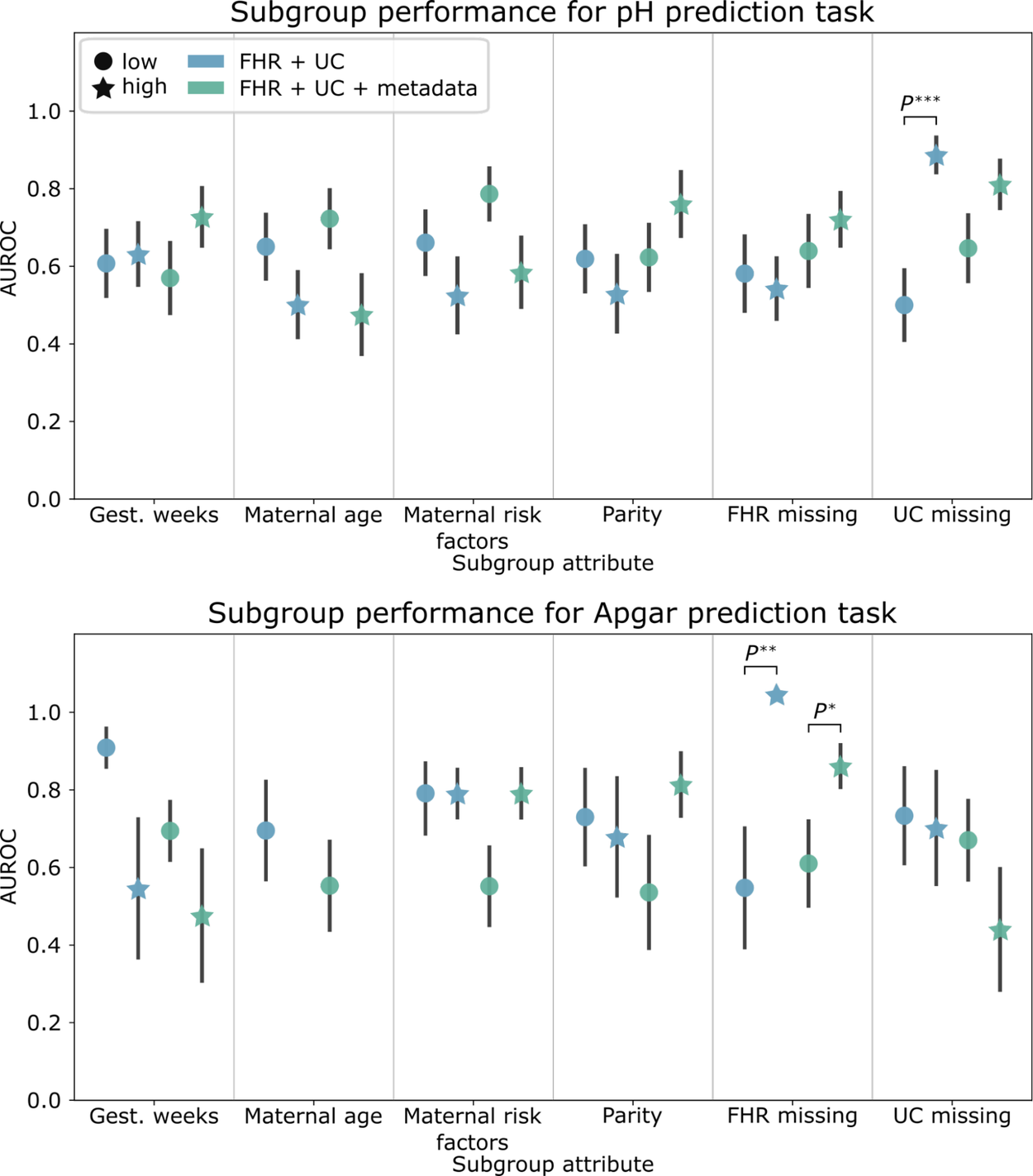
Subgroup AUROC performance for the pH (top) and Apgar (bottom) classification baseline models. Error bars depict the standard error, computed over 1,000 bootstrap samples.
An open CTG model for research use cases
We are currently exploring open-sourcing our models in hopes that other researchers and stakeholders can build on this work with their own datasets to evaluate it for their clinical use cases, keeping in mind the limitations described below.
Limitations and future work
This study had limitations that constrain the generalizability of our findings. First, we used de-identified open-source CTGs from 552 patients at a single hospital in Prague, Czech Republic. To enhance the robustness of our findings, future investigations should involve a larger and more diverse dataset sourced from maternity centers worldwide, encompassing varied clinical contexts, demographics, and outcomes. Secondly, the absence of automated CTG digitization infrastructure in many resource-limited settings necessitates the simulation of intermittent CTG use cases from facilities with digitized recordings. Additionally, our study did not include a comparison of algorithmic performance against clinicians viewing the same dataset, prompting future research to explore different human and algorithmic use combinations. Finally, further work is needed to understand how such prediction algorithms can be optimally integrated into clinical workflows to improve neonatal outcomes.
Acknowledgements
We would like to acknowledge Dr. Kwaku Asah-Opoku who inspired this work. We would also like to acknowledge Nicole Chiou who worked passionately on this project during her internship at Google, and core contributors: Mercy Asiedu, Nichole Young-Lin, Christopher Kelly, Tiya Tiyasirichokchai, Abdoulaye Diack, Julie Cattiau, Sanmi Koyejo, and Katherine Heller. Thanks to Marian Croak for her support and leadership.









