
Next generation medical image interpretation with MedGemma 1.5 and medical speech to text with MedASR
January 13, 2026
Daniel Golden, Engineering Manager, and Fereshteh Mahvar, Software Engineer, Google Research
We are updating our open MedGemma model with improved medical imaging support. We also describe MedASR, our new open medical speech-to-text model.
Quick links
The adoption of artificial intelligence in healthcare is accelerating dramatically, with the healthcare industry adopting AI at twice the rate of the broader economy. In support of this transformation, last year Google published the MedGemma collection of open medical generative AI models through our Health AI Developer Foundations (HAI-DEF) program. HAI-DEF models like MedGemma are intended as starting points for developers to evaluate and adapt to their medical use cases, and they can be easily scaled on Google Cloud through Vertex AI. The response to the MedGemma release has been incredible, with millions of downloads and hundreds of community-built variants published on Hugging Face.

Flow chart describing the intended use of MedGemma as a developer tool.
Today, we’re building on that momentum by releasing MedGemma 1.5 4B and launching the MedGemma Impact Challenge hackathon on Kaggle. Guided by direct feedback from the community, this model update enables developers to more effectively adapt MedGemma for applications that involve several medical imaging modalities:
- High-dimensional medical imaging: Computed tomography (CT), magnetic resonance imaging (MRI), and histopathology
- Longitudinal medical imaging: Chest X-ray time series review
- Anatomical localization: Localization of anatomical features in chest X-rays
- Medical document understanding: Extracting structured data from medical lab reports
MedGemma 1.5 4B also improves accuracy on core capabilities for text, medical records and 2D images over MedGemma 1 4B. We are publishing the updated 4B model size today to provide an ideal compute-efficient starting point for developers that is small enough to run offline, and developers can continue to use our MedGemma 1 27B parameter model for more complex text-based applications. Full details of the MedGemma 1.5 4B model and performance benchmarks appear in the MedGemma 1.5 model card.
We also recently released MedASR (on Hugging Face and Vertex AI), a new open automated speech recognition (ASR) model that was fine-tuned for medical dictation. The initial release of MedASR enables developers to convert medical speech to text and seamlessly pairs with MedGemma for advanced reasoning tasks.
MedGemma 1.5, MedASR and all other HAI-DEF models, like the MedSigLIP image encoder, remain free for research and commercial use and can be downloaded from Hugging Face or trained and adapted into scalable applications in the cloud on Vertex AI.
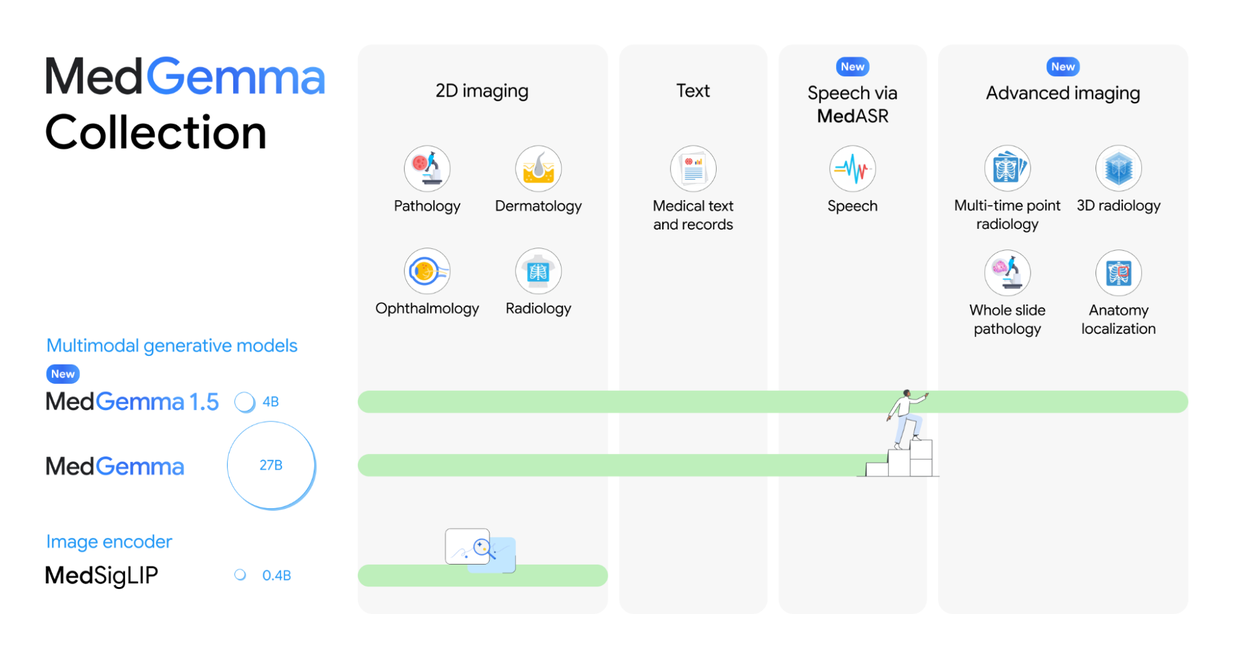
Summary of the MedGemma collection of models and their capabilities.
The MedGemma Impact Challenge
We want to encourage developers to explore additional creative and impactful uses of MedGemma models to transform healthcare. To this end, we are excited to announce the MedGemma Impact Challenge, a Kaggle-hosted hackathon with $100,000 in prizes. This hackathon is open to all developers and offers an opportunity to build on MedGemma and HAI-DEF to showcase the potential of AI in healthcare and life sciences. We look forward to what you all will build!
Improved performance for medical imaging use cases
MedGemma was designed from the ground up as a multimodal model, reflecting the multimodal nature of medicine. MedGemma 1 included support for interpreting two-dimensional medical images, including chest X-rays, dermatology images, fundus images and histopathology patches.
With MedGemma 1.5, we are expanding support for high-dimensional medical imaging, starting with three-dimensional volume representations of CT imaging and MRI, as well as whole-slide histopathology imaging. Developers can create applications in which multiple slices (for CT or MRI) or multiple patches (for histopathology) are provided as input along with a prompt that describes the task.
On internal benchmarks, the baseline absolute accuracy of MedGemma 1.5 improved by 3% over MedGemma 1 (61% vs. 58%) on classification of disease-related CT findings and by 14% (65% vs. 51%) on classification of disease-related MRI findings, averaged over findings. Additionally, on an internal diverse benchmark of histopathology slides and associated findings, the fidelity of MedGemma 1.5’s predictions, based on ROUGE-L score on cases with exactly one histopathology slide, improved by 0.47 over MedGemma 1 (0.49 vs. 0.02), matching the 0.498 score achieved by the task-specific PolyPath model.
This new high-dimensional support is the natural evolution of CT foundation, our previous API-based tool for generation of CT embeddings. To our knowledge, MedGemma 1.5 is the first public release of an open multimodal large language model that can interpret high-dimensional medical data while also retaining the ability to interpret general 2D data and text. Although these capabilities are in their early stages and remain imperfect, developers will achieve improved results by fine-tuning MedGemma models on their own data, and we hope to continually improve MedGemma models over time. We’ve released tutorial notebooks that illustrate how to use this high dimensional image capability for CT (Hugging Face, Model Garden) and histopathology (Hugging Face, Model Garden).

Example showing how MedGemma 1.5 4B can be used to interpret a CT volume along with commentary by a board-certified thoracic radiologist on the quality of the output. Note that MedGemma is not intended to be used without appropriate validation, adaptation and/or making meaningful modification by developers for their specific use case.
MedGemma 1.5 4B baseline performance also improves significantly over MedGemma 1 4B on several other forms of medical image interpretation:
- Anatomical localization: Localization of anatomical features in chest X-rays; improvement of 35% intersection over union on the Chest ImaGenome benchmark (38% vs. 3%). See our tutorial notebook on anatomical localization.
- Longitudinal medical imaging: Chest X-ray time series review; improvement of 5% macro accuracy on the MS-CXR-T benchmark (66% vs. 61%). See the example below and our tutorial notebook on longitudinal medical imaging.
- Medical image interpretation: Our internal single-image benchmarks for CXR, dermatology, histopathology and ophthalmology; improvement of 3% (62% vs. 59%).
- Lab report extraction: Extracting structured data from medical lab reports (lab type, value, units); improvement of 18% retrieval macro F1 on an internal benchmark of lab reports (78% vs. 60%).

MedGemma 1.5 4B improves support for medical imaging, exceeding the performance of MedGemma 1 4B on high-dimensional image interpretation, anatomy localization and longitudinal disease assessment in chest X-rays, general medical image interpretation and extraction of content from medical laboratory reports.

Example showing how MedGemma 1.5 4B can be used to interpret a longitudinal pair of chest X-rays along with commentary by a board-certified thoracic radiologist on the quality of the output. Note that MedGemma is not intended to be used without appropriate validation, adaptation and/or making meaningful modification by developers for their specific use case.
Additionally, MedGemma applications that are deployed on Google Cloud now include full DICOM support, making it even easier to adapt MedGemma for medical imaging applications.
Improvements to text functionality
Beyond the improved support for medical images, we’ve worked hard to improve the baseline medical text functionality of MedGemma. Through the addition of new training datasets and training techniques, MedGemma 1.5 4B improves over MedGemma 1 4B by 5% on MedQA (69% vs. 64%) and by 22% on text-based EHR question-answering with EHRQA (90% vs. 68%).

MedGemma 1.5 4B improves at text-based tasks over MedGemma 1 4B, including on medical reasoning (MedQA) and electronic health record information retrieval (EHRQA).
MedASR: An open model for medical automated speech recognition
While text is currently the primary interface for large language models, verbal communication remains crucial in many aspects of healthcare, including medical dictation and live conversations between patients and providers. Speech also provides a more natural way to interact with a language model.
To support these use cases that require the model to be familiar with healthcare’s specialized vocabulary, we developed the MedASR speech to text model to transcribe speech from the medical domain. MedASR can be used both to transcribe medical dictation and as a natural method of generating prompts for MedGemma. We compared MedASR’s performance to Whisper large-v3, a generalist ASR model, and found that MedASR had 58% fewer errors on chest X-ray dictations (5.2% vs. 12.5% word error rate, WER) and 82% fewer errors on an internal medical dictation benchmark with diverse specialties and speakers (5.2% vs. 28.2% WER). We’ve released a collection of tutorial notebooks to help developers create and adapt their own systems that combine the audio understanding of MedASR with the clinical reasoning of MedGemma 1.5. Learn more in the MedASR model card.

MedASR can be used either for transcribing medical dictation (top) or to dictate prompts for MedGemma (bottom).
How developers are using MedGemma
We’re seeing health tech startups and developers around the world leverage MedGemma to accelerate their research and product development across a broad range of use cases and settings.
As one example, Qmed Asia has adapted MedGemma for use into askCPG, a conversational interface to Malaysia’s 150+ clinical practice guidelines. According to the Ministry of Health Malaysia, the conversational interface has made navigating the Malaysian Clinical Practice Guidelines more practical in day-to-day clinical decision support, and the multimodal medical image extension with MedGemma has been especially well-received in pilot deployments.
Additionally, Taiwan’s National Health Insurance Administration has applied MedGemma for evaluating preoperative assessments for lung cancer surgery. By extracting key data from over 30,000 pathology reports and unstructured data using MedGemma, they performed statistical analyses to assess the preoperative medical condition of patients. This effort aims to inform policy decisions that improve decision-making for surgical resection in order to improve patient outcomes.
MedGemma has also been cited extensively in medical AI research articles since its release earlier this year, comparing favorably to other models as a base model for understanding medical text, multidisciplinary team decision making, mammography reporting, and other clinical scenarios.
Get started
You can access all variants of MedGemma via the Hugging Face collection or Vertex AI on Google Cloud. MedASR is currently available on Hugging Face and Vertex AI. To showcase your ideas for the next generation of medical AI applications, check out the MedGemma Impact Challenge.
Visit our MedGemma GitHub repository to explore our expanded collection of tutorials. These include our existing tutorials on running inference and LoRA-based supervised fine-tuning and a new tutorial on reinforcement learning, a tuning method that is particularly effective at learning complex tasks without compromising existing model abilities.
Visit the HAI-DEF site for resources on MedGemma 1.5 and other Health AI Developer Foundations models. To stay up to date, please sign up for our newsletter. For technical support, please use the HAI-DEF forum.
We’re very excited by what the community will build with these new models and welcome your feedback.

This table summarizes model features and can help you understand which model is ideal for your use case.
Note on datasets
Models were trained and evaluated on a mix of public and private de-identified datasets. Google and its partners utilize datasets that have been rigorously anonymized or de-identified to ensure the protection of individual research participants and patient privacy.
Disclaimer
HAI-DEF models, including MedGemma and MedASR, are intended to be used as a starting point that enables efficient development of downstream healthcare applications involving medical text and images. HAI-DEF models are not intended to be used without appropriate validation, adaptation and/or making meaningful modification by developers for their specific use case. The outputs generated by these models are not intended to directly inform clinical diagnosis, patient management decisions, treatment recommendations, or any other direct clinical practice applications. Performance benchmarks reported here highlight baseline capabilities and are not intended to imply that MedGemma is safe to use in any given medical application. Inaccurate model outputs beyond those shown here are possible. All model outputs should be considered preliminary and require independent verification, clinical correlation, and further investigation through established research and development methodologies. Please refer to terms of use and prohibited use policy for more details.
Acknowledgments
MedGemma, the MedGemma Impact Challenge, and MedASR are the products of a collaboration between teams at Google. We thank the many people who contributed to this work, including the engineering and cross-functional members of the Health AI, Gemma, and Kaggle teams, as well as our sponsors in Google Research and Google DeepMind.










