Insulin resistance prediction from wearables and routine blood biomarkers
August 6, 2025
Ahmed A. Metwally, Staff Research Scientist, and A. Ali Heydari, Research Scientist, Google Research
Leveraging wearable data and routine blood tests, we propose a novel method for effectively predicting insulin resistance, providing a scalable and accessible approach for early type 2 diabetes risk screening.
Quick links
Type 2 diabetes affects hundreds of millions globally, and its prevalence is rising. A major precursor to this condition is insulin resistance (IR), where the body's cells do not respond properly to insulin, a hormone crucial for regulating blood sugar. Detecting IR early is key, as lifestyle changes can often reverse it and prevent or delay the onset of type 2 diabetes. However, current methods for accurately measuring IR, like the "gold standard" euglycemic insulin clamp or the Homeostatic Model Assessment for Insulin Resistance (HOMA-IR), which requires specific insulin blood tests, are often invasive, expensive, or not readily available in routine check-ups. These steps create significant barriers to early detection and intervention, especially for those unknowingly at risk.
What if we could leverage data already available to many people, such as data from wearable devices and common blood tests, to estimate IR risk? In “Insulin Resistance Prediction From Wearables and Routine Blood Biomarkers”, we explore a suite of machine learning models that have the potential of predicting IR using wearable data (e.g., resting heart rate, step count, sleep patterns) and routine blood tests (e.g., fasting glucose, lipid panel). This approach shows strong performance across the studied population (N=1,165) and an independent validation cohort (N=72), particularly in high-risk individuals, such as people with obesity and sedentary lifestyles. Additionally, we introduce the Insulin Resistance Literacy and Understanding Agent (an IR prototype agent), built on the state-of-the-art Gemini family of LLMs to help understand insulin resistance, facilitating interpretation and safe personalized recommendations. This work offers the potential for early detection of people at risk of type 2 diabetes and thereby facilitates earlier implementation of preventative strategies. The models, predictions, and the Insulin Resistance Literacy and Understanding Agent described in this research are intended for informational and research purposes only.

Metabolic subphenotypes of type 2 diabetes. Chronic insulin resistance is a precursor to approximately 70% of type 2 diabetes cases and results from a combination of obesity, an inactive lifestyle, and genetic factors.
Predicting IR using digital biomarkers & routine blood tests
We designed a study, called WEAR-ME, to explore the potential of predicting insulin resistance (through predicting HOMA-IR) using readily accessible data. To automate the data collection process for routine blood biomarkers, we partnered with Quest Diagnostics.
1,165 remote participants from across the US signed up for the WEAR-ME study via the Google Health Studies app, a secure consumer-facing platform for digital studies. This study was conducted with approval from an Institutional Review Board (IRB). All participants provided electronic informed consent and a specific HIPAA Authorization via the Google Health Studies app before enrollment. The cohort was diverse in age, gender, geography, and BMI. Participants had a median BMI of 28 kg/m², age of 45 years, and HbA1c of 5.4%. Participants consented to share the following data:
- Wearable Data: Data from their Fitbit or Google Pixel Watch devices (like resting heart rate, step count, sleep patterns), pseudonymized to protect individual participant privacy.
- Routine Blood Biomarkers: Results from routine tests (like fasting glucose and insulin, lipid panel) ordered during an in-person visit to Quest Diagnostics, specifically for this research.
- Demographics and Surveys: Basic information and health questionnaires (completed at the beginning and end of the study), which included data on age, weight, height, ethnicity, race, and gender, along with questions about perception of overall health (fitness, diet) and history of diabetes or other comorbidities.
Using this rich, multimodal dataset (which we refer to as the “WEAR-ME data”), we developed and trained deep neural network models to predict HOMA-IR scores. Our goal was to see how well we could estimate this key IR marker using different combinations of available data.

Illustration of our proposed modeling pipeline for predicting HOMA-IR, and interpreting the results with the Insulin Resistance Education and Understanding Agent.
Our results, using the area under the receiver operating characteristic curve (auROC) metric, indicate that combining data streams significantly improved prediction accuracy compared to using any single source alone:
- Wearables + Demographics: Showed some predictive power (auROC = 0.70) for classifying IR.
- Adding Fasting Glucose to Wearables + Demographics: This routine blood test result proved highly valuable, significantly boosting performance (auROC = 0.78).
Wearables + Demographics + Routine Blood Panels: Achieved the best results, accurately predicting HOMA-IR values (R² = 0.50) and effectively classifying individuals with IR (auROC = 0.80, Sensitivity = 76%, Specificity = 84%, where HOMA-IR value of 2.9 or higher was used to identify a person as being insulin resistant).

Left: Performance evaluation of IR prediction (classification). Right: Visualization of the precision-recall curve for selected feature sets. Average values are colors, with the gray areas around each line indicate the standard deviation across the five folds.
Importantly, our results indicate that features derived from wearable data, such as resting heart rate, consistently ranked among the most important predictors, alongside BMI and fasting glucose. The feature importance result highlights the value of capturing lifestyle-related signals.
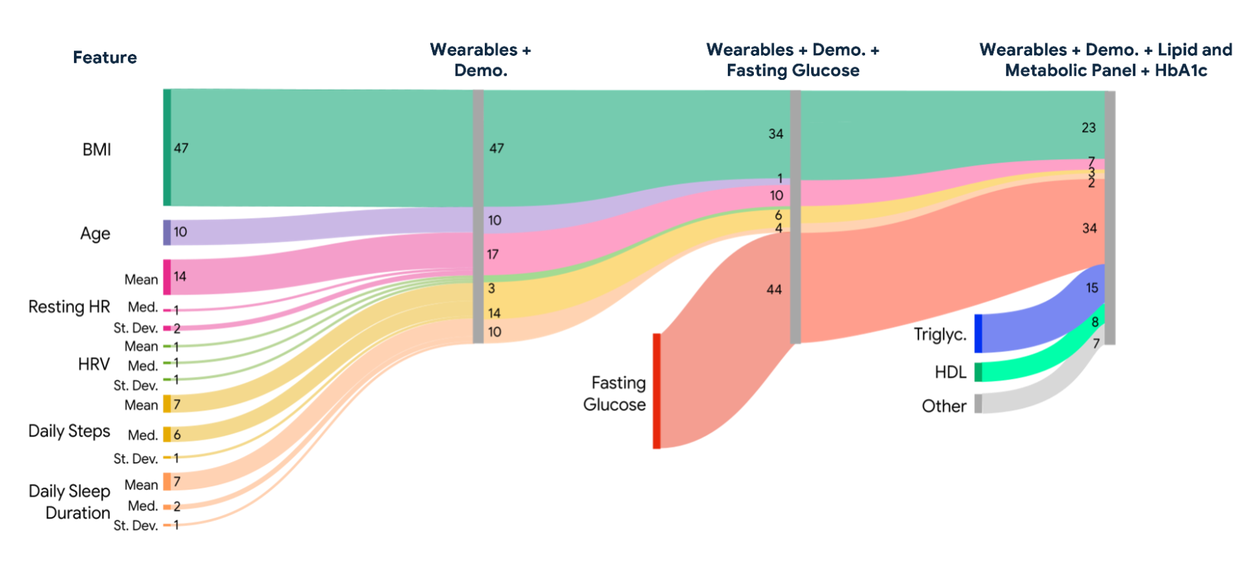
Sankey diagram showing the relative feature importance (SHapley Additive exPlanations [SHAP] values) for each of the proposed nonlinear XGBoost models for direct regression.
Focusing on high-risk groups
Since individuals with obesity and sedentary lifestyles are particularly vulnerable to developing type 2 diabetes, we specifically evaluated our model's performance in these subgroups:
- Obese Participants: The model showed improved accuracy, compared to the overall population (sensitivity = 86% vs %76).
- Sedentary Participants: Accuracy was higher than the obese subpopulation (sensitivity = 88%).
- Obese and Sedentary Participants: The model performed notably well in this critical group (sensitivity = 93%, adjusted specificity = 95%; adjusted specificity here focuses on minimizing the misclassification of truly insulin-sensitive individuals as resistant).
The results of this experiment suggest that our approach could be particularly effective at identifying those who might benefit most from early lifestyle interventions.

Results of classification performance for various lifestyle stratification.
Validation and generalizability
To ensure our findings were not just specific to our initial dataset, we tested our best-performing model (trained on the WEAR-ME data) on a completely independent validation cohort (N=72) recruited through a separate IRB-approved consented study, where participants shared wearable data using the Fitbit Charge 6, and blood biomarker data was acquired in-person at the study center in San Francisco. This cohort had a median BMI of 30.6 kg/m² and age of 44.5 years. Our results on the validation cohort show that our trained models maintained strong predictive performance (sensitivity = 84%, specificity = 81%), demonstrating its potential generalizability. However, as this remains a research prototype, its safety and effectiveness for any health-related purpose have not been established.

Overview of the independent validation cohort study. We compare model accuracies from the initial training and testing cohort with the external validation cohort and demonstrate its potential generalizability.
Beyond prediction: Towards understanding and proactive steps
Illustration of the proposed agentic architecture that leverages the HOMA-IR prediction model to assess insulin resistance risk to educate users.
Predicting IR risk is valuable, but how can we make this information understandable and actionable for individuals? We explored integrating our prediction models with LLMs to empower users to better understand their metabolic health. We developed the Insulin Resistance Literacy and Understanding Agent (an IR prototype agent), built on the state-of-the-art Gemini family of LLMs. When asked a question about metabolic health, the IR Agent provides personalized, contextualized answers for educational purposes grounded in the individual's study data and predicted IR status. With the user's consent, the agent has the ability to access specific, user-provided data points, search for up-to-date information, and perform calculations. It is critical to note that interaction with the models or the IR Agent are intended to demonstrate how such a tool could help users explore their results for informational and educational purposes.
We had five board-certified endocrinologists evaluate responses from the IR Agent compared to a base model. They strongly preferred the IR Agent's responses, finding them to be significantly more comprehensive, trustworthy, and personalized. This demonstrates the potential of combining predictive health models with LLMs to empower individuals with better health understanding.

Overview of Insulin Resistance Literacy and Understanding Agent (IR Agent). An illustration of the proposed IR agent (left), along with the results (win rate) of our IR agent against the base model as evaluated by endocrinologists (right).
Conclusions and future work
Our research demonstrates that ML models combining readily available wearable data and routine blood biomarkers have the potential to effectively predict insulin resistance, a key precursor to type 2 diabetes. This approach offers several advantages:
- Accessibility: Leverages data many people already have or can easily obtain.
- Early Detection: Identifies risk even before blood sugar levels become abnormal, e.g., we found many normoglycemic (with HbA1c < 5.7) participants in our study already had IR.
- Scalability: Offers a potentially more scalable screening method than specialized IR tests.
- Personalization: Shows strong performance in high-risk subgroups and potential for integration into personalized health tools.
This work opens doors for earlier, more accessible screening of type 2 diabetes risk, potentially enabling timely lifestyle interventions that could prevent or delay the disease, particularly for those unknowingly progressing towards it.
Future work includes validating these models longitudinally (tracking individuals over time), exploring the impact of interventions, incorporating genetic and microbiome data, and further refining models for specific populations to ensure equitable performance across diverse groups. We believe this line of research holds significant promise for proactive and personalized metabolic health management.
Disclaimer
While our proposed approach, including the IR Agent, holds promise for various health applications, this research specifically addresses the critical need for early detection of insulin resistance, and does not present the models discussed herein as approved medical devices or solutions. The models and the IR Agent are not medical devices. They have not been cleared, approved, or reviewed by the U.S. Food and Drug Administration (FDA) or any other national or international regulatory agency. This work is not intended to be, and should not be used as, a substitute for professional medical advice, diagnosis, or treatment. Real-world deployment of such technologies would necessitate rigorous testing, validation, and regulatory approval.
Acknowledgements
The research described here is joint work across Google Research and partnering teams. The following researchers contributed to this work: Ahmed A. Metwally, A. Ali Heydari, Daniel McDuff, Alexandru Solot, Zeinab Esmaeilpour, Anthony Z. Faranesh, Menglian Zhou, David B. Savage, Conor Heneghan, Shwetak Patel, Cathy Speed, and Javier L. Prieto. Google partnered with Quest Diagnostics, the world’s leading provider of diagnostic information, to allow eligible participants to share their biomarker data received as part of a free blood draw, which includes a comprehensive metabolic panel and measuring cholesterol, triglycerides and insulin levels.









