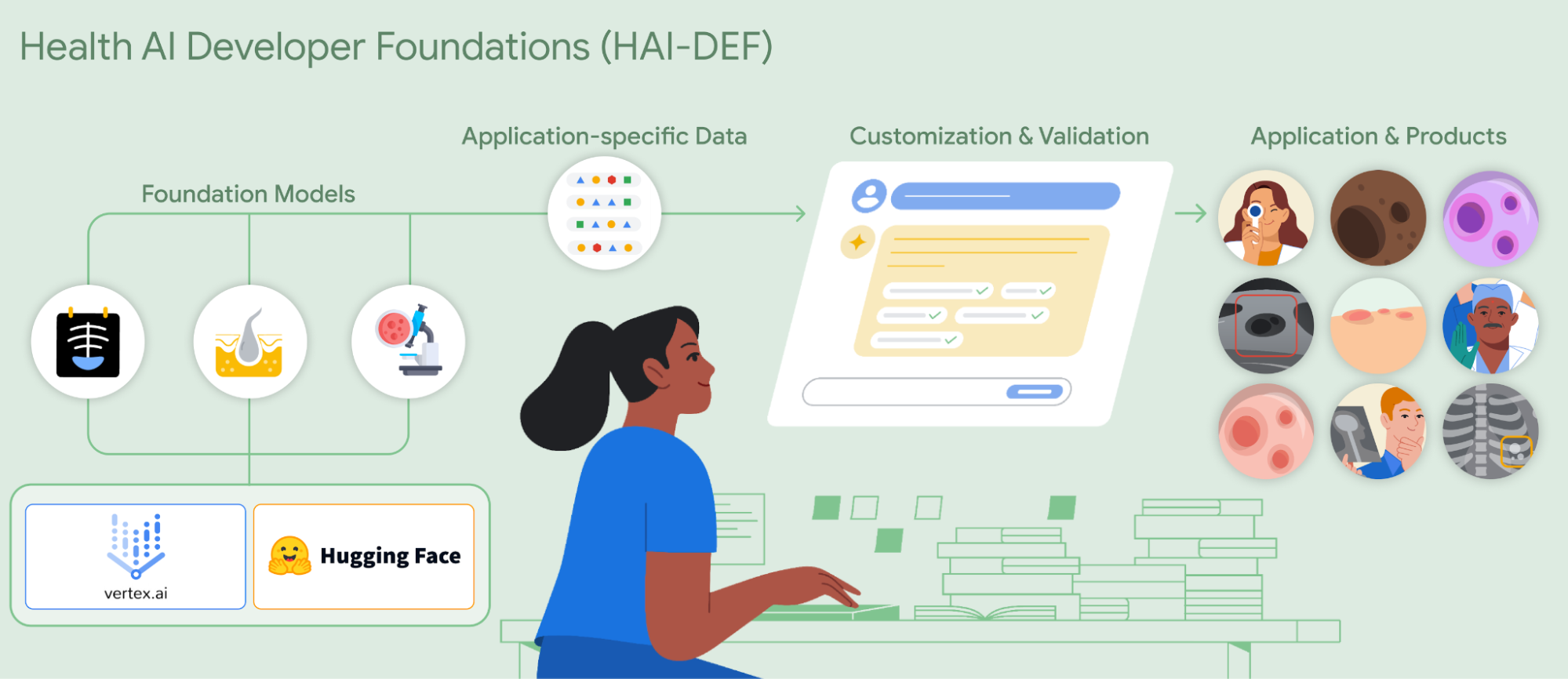
Helping everyone build AI for healthcare applications with open foundation models
November 25, 2024
Tim Thelin and Can Kirmizibayrak, Software Engineers, Google Research, Health AI Team
Health AI Developer Foundations (HAI-DEF) is a new suite of open weight models to help developers more easily build AI models for healthcare applications. The initial launch is focused on imaging applications in radiology, dermatology and pathology.
Quick links
AI can have a tremendous potential impact on healthcare by helping to improve diagnostic accuracy, broadening access to care, and easing administrative burden to enable care teams to focus on their patients. However, the field of healthcare is vast and there are more potential use-cases than developers can cover. In addition, AI development for health is particularly challenging because of the amount of data, expertise, and compute required to build models that reach the performance levels necessary for use in a clinical setting.
Without sufficiently diverse data — e.g., across patient populations, data acquisition devices, or protocols — models may not generalize well when deployed in environments that differ from the data on which they were trained. The resulting high barrier to entry prevents many would-be health AI developers from experimenting and makes it more difficult for them to take their ideas from concept to prototype, much less bench to bedside. For healthcare to continue to realize its potential, it needs innovation from a diverse set of contributors on a multitude of use-cases, interfaces and business models.
With this in mind, today we're introducing Health AI Developer Foundations (HAI-DEF), a public resource to help developers build and implement AI models for healthcare more efficiently. Summarized in an accompanying technical report, HAI-DEF includes open-weight models, instructional Colab notebooks, and documentation to assist in every stage of development, from early research to commercial ventures.
HAI-DEF is part of our broader commitment to support healthcare AI development. It builds upon the Medical AI Research Foundations repository, released in 2023, which includes models for chest X-ray and pathology images. It also compliments initiatives like Open Health Stack, also launched in 2023, which provides developers with open-source building blocks for building effective health apps, and Population Dynamics Foundation Model, launched in 2024, which provides developers with geospatial embeddings to enable modeling of population-level changes including public health and beyond. By providing resources such as these, we aim to democratize AI development for healthcare, empowering developers to create innovative solutions that can improve patient care.
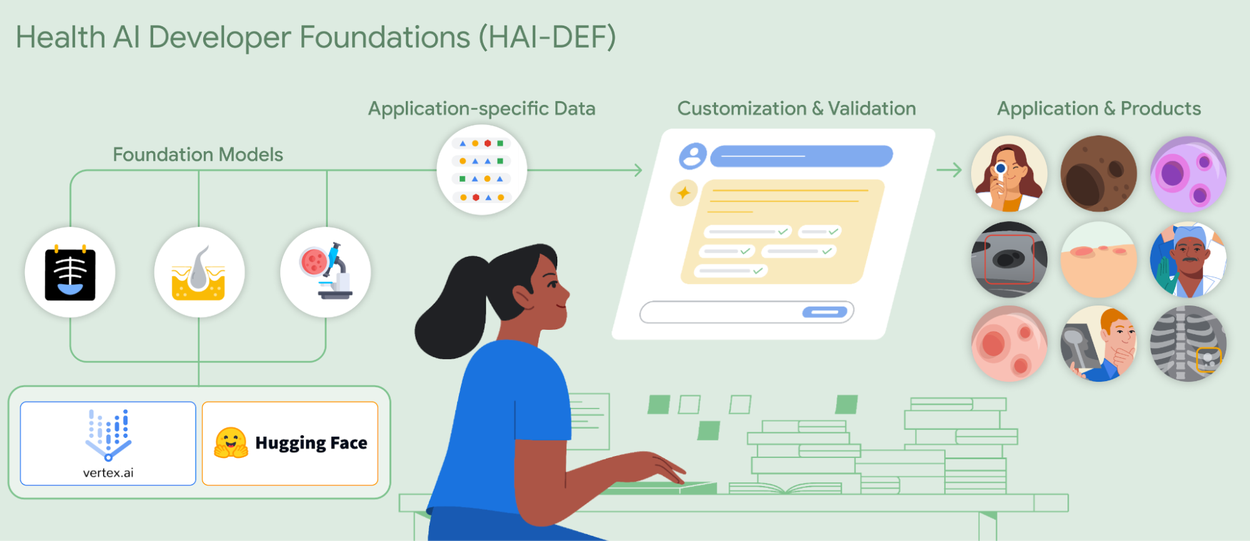
HAI-DEF’s inaugural models
The inaugural release of HAI-DEF includes three models focused on supporting development of medical imaging applications:
- CXR Foundation for chest X-rays
- Derm Foundation for skin images
- Path Foundation for digital pathology
Each of these is an embedding model specialized for a specific medical imaging modality. They improve the efficiency of training and serving models, taking images as inputs and producing fixed-length vectors (embeddings) that efficiently represent the input image. The models are developed from extensive, self-supervised training on large amounts of diverse, de-identified data for their respective modalities. As a result, the embeddings the models produce provide a powerful starting point for developers to build high performing AI models for their own use cases, with a very small amount of additional data and compute.
CXR Foundation is pre-trained using EfficientNet-L2 architecture on over 800,000 X-rays. It was trained using Supervised Contrastive, CLIP and BLIP-2 losses. As part of its BLIP training phase it also features a BERT-based text encoder that allows it to project text and images into a shared embedding space. CXR Foundation’s image encoding model takes DICOM images, and its text-encoder accepts textual strings. This allows users to do data-efficient classification, building small models on top of the embeddings to classify conditions the user cares about. The language component allows the user to also do:
- Semantic image search: ranking a set of images by their closeness in embedding space to some search term; and
- Zero-shot classification: using the distance between textual terms and the image embedding to provide a classification score with no example images required. Note the performance of zero-shot will be lower than data-efficient classification.
Derm Foundation is based on the BiT ResNet-101x3 architecture. It was pre-trained on a wide range of skin images to produce enriched embeddings useful for data-efficient classification of skin related tasks. These could include clinical tasks, such as dermatitis, melanoma or psoriasis but could also be used for understanding which body part(s) are involved, determining image quality, and whether the photograph should be retaken.
Path Foundation is an efficient embedding model, trained from a ViT-S architecture, specialized on hematoxylin and eosin (H&E) stained images. Path Foundation accepts 224 x 224 pixel patches from H&E slides to produce embeddings that can be used for data-efficient classification for applications like grading or identifying tumors, classifying tissue or stain type, and determining image quality. The embeddings can also be used for similar image search tasks, to find areas within or across slides that resemble each other.
Learning from community experience with previous research endpoints
Over the last 2 years, researchers across academia, healthcare institutions, and pharma companies have been building with these models through a Google Research–hosted API. After giving the community time to use the models and explore different applications, we collected feedback. Many desired to download the models directly to enable use with datasets that cannot leave institutional boundaries. In addition, users who saw the potential of foundation models to improve clinical workflows wanted to build towards use cases with an eye towards helping clinical care.
In response to this feedback, HAI-DEF will enable developers to:
- Download and run these models in their own environment whether locally or on the cloud;
- Use them to develop applications for research or commercial ventures; and
- Fine-tune them to achieve even better performance.
The models are accessible via Vertex AI Model Garden [CXR, Derm, Path] and Hugging Face [CXR, Derm, Path]. Because the model weights are open, developers can fine tune the models to improve performance for their specific needs and applications, use the embedding models as part of complex ensembles or hybrid architectures, and more.
Building a health AI developer ecosystem
HAI-DEF is just one of the ways we're enabling the broader ecosystem to build for health, supplementing Open Health Stack and Population Dynamics Foundation Model. We are excited to continue investing in this space, including by adding more models to HAI-DEF and expanding the scope of our notebooks. We look forward to seeing the community build on these resources to realize AI’s potential to transform healthcare and life sciences.
Acknowledgements
We thank Google Health team members who led this research and made the public release possible, including Rory Pilgrim, Fereshteh Mahvar, Liron Yatziv, Tiffany Chen, Kenneth Philbrick, Bram Sterling, Nick George, Atilla Kiraly, Sebastien Baur, Fayaz Jamil, Bill Luan, Howard Yang, Preeti Singh, Faruk Ahmed, Lin Yang, Andrew Sellergren, Daniel Golden, Abbi Ward, Shruthi Prabhakara, Jennifer Klein, Chuck Lau, Jason Klotzer, Shekoofeh Azizi, Rachelle Sico, Anthony Phalen, Amanda Ferber, Lauren Winer, Jenn Sturgeon, David F. Steiner, Yun Liu and Shravya Shetty. Credits to Tiya Tiyasirichokchai for creation of the figure.