An NLU-Powered Tool to Explore COVID-19 Scientific Literature
May 4, 2020
Posted by Keith Hall, Research Scientist, Natural Language Understanding, Google Research
Quick links
Update — 2021/05/20: We are expanding the Research Explorer to include all PubMed publications, a corpus of over 31 million scientific papers, as well as content from PubMed Central. We’re using similar technology as described in the post below, so if you’re curious about how the NLP models work, please read on. Additionally, we have added “entity” labels under each search result; these are key words or phrases which have been identified in the returned snippets. Finally, you can now export your search results to a CSV file or directly to Google Sheets. Try the expanded BioMed Explorer at g.co/research/biomedexplorer.
Due to the COVID-19 pandemic, scientists and researchers around the world are publishing an immense amount of new research in order to understand and combat the disease. While the volume of research is very encouraging, it can be difficult for scientists and researchers to keep up with the rapid pace of new publications. Traditional search engines can be excellent resources for finding real-time information on general COVID-19 questions like "How many COVID-19 cases are there in the United States?", but can struggle with understanding the meaning behind research-driven queries. Furthermore, searching through the existing corpus of COVID-19 scientific literature with traditional keyword-based approaches can make it difficult to pinpoint relevant evidence for complex queries.
To help address this problem, we are launching the COVID-19 Research Explorer, a semantic search interface on top of the COVID-19 Open Research Dataset (CORD-19), which includes more than 50,000 journal articles and preprints. We have designed the tool with the goal of helping scientists and researchers efficiently pore through articles for answers or evidence to COVID-19-related questions.
When the user asks an initial question, the tool not only returns a set of papers (like in a traditional search) but also highlights snippets from the paper that are possible answers to the question. The user can review the snippets and quickly make a decision on whether or not that paper is worth further reading. If the user is satisfied with the initial set of papers and snippets, we have added functionality to pose follow-up questions, which act as new queries for the original set of retrieved articles. Take a look at the animation below to see an example of a query and a corresponding follow-up question. We hope these features will foster knowledge exploration and efficient gathering of evidence for scientific hypotheses.
Semantic Search
A key technology powering the tool is semantic search. Semantic search aims to not just capture term overlap between a query and a document, but to really understand whether the meaning of a phrase is relevant to the user’s true intent behind their query.
Consider the query, “What regulates ACE2 expression?” Even though this seems like a simple question, certain phrases can still confuse a search engine that relies solely on text matching. For example, “regulates” can refer to a number of biological processes. While traditional information retrieval (IR) systems use techniques like query expansion to mitigate this confusion, semantic search models aim to learn these relationships implicitly.
Word order also matters. ACE2 (angiotensin converting enzyme-2) itself regulates certain biological processes, but the question is actually asking what regulates ACE2. Matching on terms alone will not distinguish between “what regulates ACE2 ” and “what ACE2 regulates.” Traditional IR systems use tricks like n-gram term matching, but semantic search methods strive to model word order and semantics at their core.
The semantic search technology we use is powered by BERT, which has recently been deployed to improve retrieval quality of Google Search. For the COVID-19 Research Explorer we faced the challenge that biomedical literature uses a language that is very different from the kinds of queries submitted to Google.com. In order to train BERT models, we required supervision — examples of queries and their relevant documents and snippets. While we relied on excellent resources produced by BioASQ for fine-tuning, such human-curated datasets tend to be small. Neural semantic search models require large amounts of training data. To augment small human-constructed datasets, we used advances in query generation to build a large synthetic corpus of questions and relevant documents in the biomedical domain.
Specifically, we used large amounts of general domain question-answer pairs to train an encoder-decoder model (part a in the figure below). This kind of neural architecture is used in tasks like machine translation that encodes one piece of text (e.g., an English sentence) and produces another piece of text (e.g., a French sentence). Here we trained the model to translate from answer passages to questions (or queries) about that passage. Next we took passages from every document in the collection, in this case CORD-19, and generated corresponding queries (part b). We then used these synthetic query-passage pairs as supervision to train our neural retrieval model (part c).
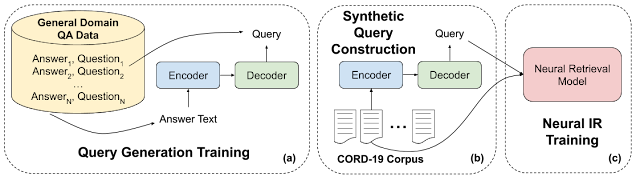 |
| Synthetic query construction. |
However, we found that there were examples where the neural model performed worse than a keyword-based model. This is because of the memorization-generalization continuum, which is well known in most fields of artificial intelligence and psycholinguistics. Keyword-based models, like tf-idf, are essentially memorizers. They memorize terms from the query and look for documents that have them. Neural retrieval models, on the other hand, learn generalizations about concepts and meaning and try to match based on those. Sometimes they can over-generalize when precision is important. For example, if I query, “What regulates ACE2 expression?”, one may want the model to generalize the concept of “regulation,” but not ACE2 beyond acronym expansion.
Hybrid Term-Neural Retrieval Model
To improve our system we built a hybrid term-neural retrieval model. A crucial observation is that both term-based and neural models can be cast as a vector space model. In other words, we can encode both the query and documents and then treat retrieval as looking for the document vectors that are most similar to the query vector, also known as k-nearest neighbor retrieval. There is a lot of research and engineering that is needed to make this work at scale, but it allows us a simple mechanism to combine methods. The simplest approach is to combine the vectors with a trade-off parameter.
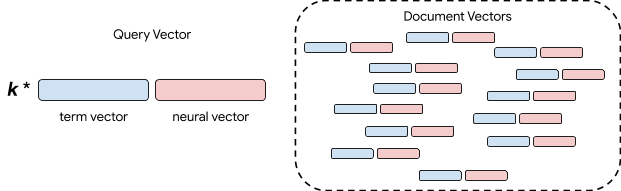 |
| Hybrid Term and Neural Retrieval. |
In the figure above, the blue boxes are the term-based vectors, and the red, the neural vectors. We represent documents by concatenating these vectors. We concatenate the two vectors for queries as well, but we control the relative importance of exact term matches versus neural semantic matching. This is done via a weight parameter k. While more complex hybrid schemes are possible, we found that this simple hybrid model significantly increased quality on our biomedical literature retrieval benchmarks.
Availability and Community Feedback
The COVID-19 Research Explorer is freely available to the research community as an open alpha. Over the coming months we will be making a number of usability enhancements, so please check back often. Try out the COVID-19 Research Explorer, and please share any comments you have with us via the feedback channels on the site.
Acknowledgements
This effort has been successful thanks to the hard work of many people, including, but not limited to the following (in alphabetical order of last name): John Alex, Waleed Ammar, Greg Billock, Yale Cong, Ali Elkahky, Daniel Francisco, Stephen Greco, Stefan Hosein, Johanna Katz, Gyorgy Kiss, Margarita Kopniczky, Ivan Korotkov, Dominic Leung, Daphne Luong, Ji Ma, Ryan Mcdonald, Matt Pearson-Beck, Biao She, Jonathan Sheffi, Kester Tong, Ben Wedin
-
Labels:
- Natural Language Processing