Accelerating scientific discovery with AI-powered empirical software
September 9, 2025
Lizzie Dorfman, Product Manager, and Michael Brenner, Research Scientist, Google Research
Our new AI system helps scientists write empirical software, achieving expert-level results on six diverse, challenging problems.
Quick links
In scientific research, thoroughly evaluating hypotheses is essential to developing more robust and comprehensive answers, but the required work forms a bottleneck, hindering the pace of discovery. In particular, much of modern scientific research depends on computational experiments to model, simulate, and analyze complex phenomena. Here, hypothesis evaluation often requires creating custom software, a slow and challenging task. Given the increasing capability of large language models (LLMs) to perform traditional coding tasks, we wondered if they could similarly generate high-quality custom software for evaluating and iteratively improving scientific hypotheses.
Today we are releasing a paper describing an "AI system designed to help scientists write expert-level empirical software", built using Gemini. Taking as input a well-defined problem and a means of evaluation, our system acts as a systematic code-optimizing research engine: it can propose novel methodological and architectural concepts, implement them as executable code and empirically validate their performance. It then searches and iterates through thousands of code variants, using tree search to optimize performance. We tested our system using six benchmarks representing distinct multidisciplinary challenges, spanning the fields of genomics, public health, geospatial analysis, neuroscience, time-series forecasting, and numerical analysis. Our system achieves expert-level performance across all of these benchmarks.
Empirical software and scorable tasks
Scientific research is inherently iterative, often requiring researchers to test dozens or hundreds of models or parameters to achieve a breakthrough. Even for scientists who are experienced programmers, coding, debugging, and optimizing software is incredibly time-consuming. Manually coding each new idea is slow and inefficient, making systematic exploration of potential solutions practically impossible.
At the heart of our system lies the foundational concept of empirical software. Unlike conventional software, which is often judged by functional correctness alone, empirical software is designed with a primary objective: to maximize a predefined quality score. A problem or challenge that can be effectively addressed and solved through the application of empirical software is termed a scorable task. These scorable tasks are prevalent across science, applied mathematics, and engineering.
How it works
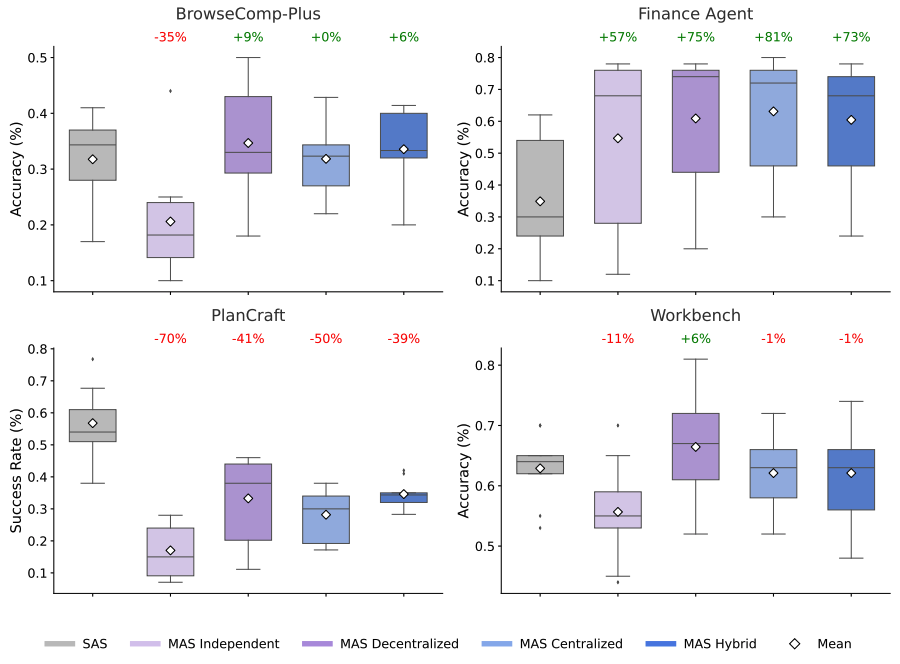
The input to our system is a scorable task, which includes a problem description, a scoring metric, and data suitable for training, validation, and evaluation. A user can also provide context, such as ideas from external literature, or directives for methodologies to prioritize.
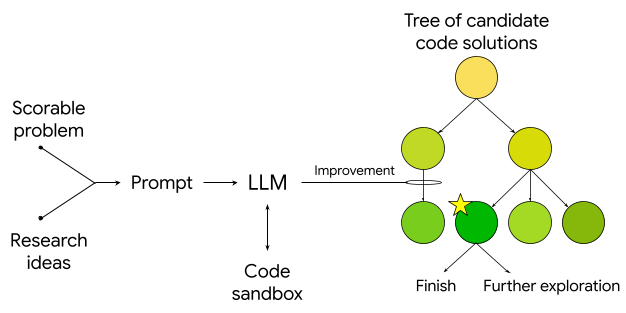
The system then generates research ideas, including programmatic reproduction, optimization, and recombination of known methods, leading to novel and highly performant approaches. Ideas are implemented as executable code and the system uses a tree search strategy with an upper confidence bound (inspired by AlphaZero) to create a tree of software candidates and decide which candidates warrant further exploration. It then uses an LLM to rewrite the code to attempt to improve its quality score, and can exhaustively and tirelessly carry out solution searches at an unprecedented scale, identifying high-quality solutions quickly, reducing exploration time from months to hours or days. Its outputs, as coded solutions, are verifiable, interpretable and reproducible.
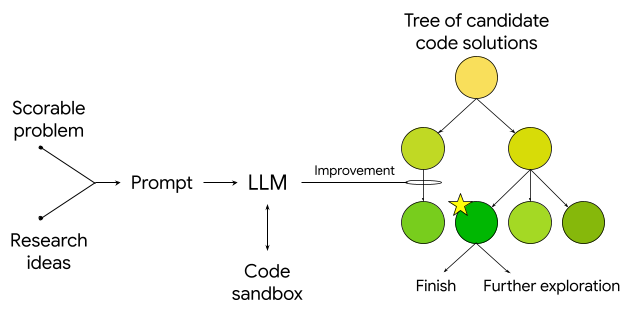
Schematic of the algorithm that feeds a scorable task and research ideas to an LLM, which generates evaluation code in a sandbox. This code is then used in a tree search, where new nodes are created and iteratively improved using the LLM.
Demonstrated effectiveness
The evaluation of code generating AI systems has historically focused on tasks derived from competitive programming or software engineering, which, while valuable, fail to capture the full spectrum of challenges inherent in scientific discovery. We demonstrate proficiency not merely in writing syntactically correct code, but in generating novel solutions to six diverse and challenging benchmark problems that push the boundaries of current computational methods and human expertise. The diversity of these benchmarks allows us to collectively assess proficiency in areas such as zero-shot generalization, high-dimensional signal processing, uncertainty quantification, semantic interpretation of complex data, and systems-level modeling. The top scoring solutions to each of these benchmark problems are openly available for anyone interested in reproducing our results, including as an interactive website to explore the full candidate solution trees.
Genomics: Batch integration of single cell RNA sequencing data
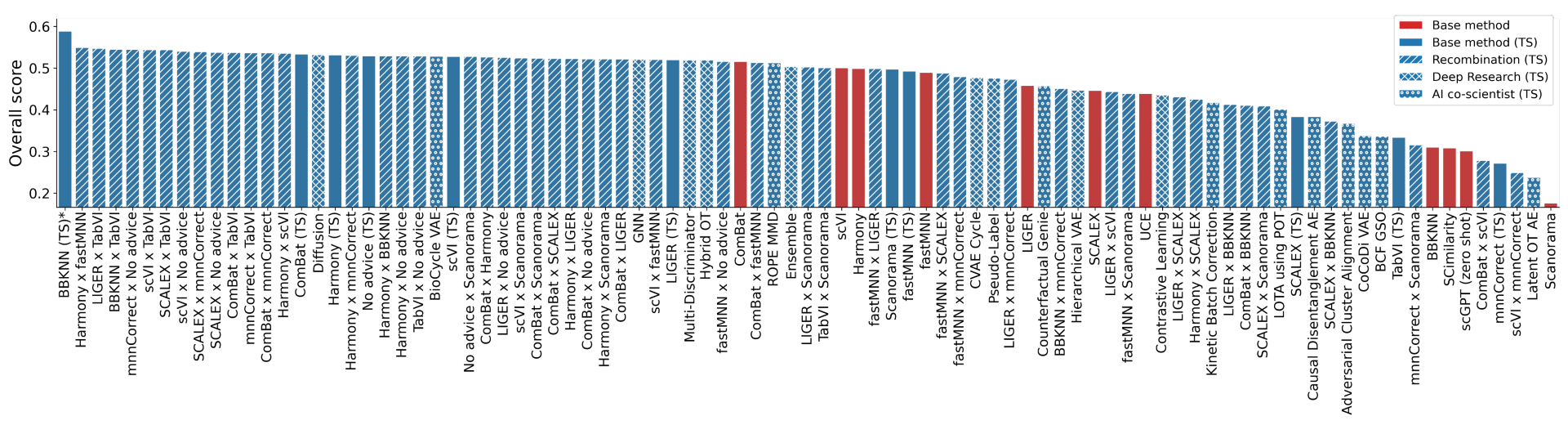
Single-cell RNA sequencing (scRNA-seq) is a powerful technology that provides a high-resolution view of gene expression at the individual cell level. A major challenge required to jointly analyze many disparate datasets is to remove complex batch effects present across samples while preserving true biological signals. Nearly 300 tools exist to perform batch integration of scRNA-seq data, and multiple benchmarks have been developed for assessing metrics of batch effect removal and conservation of biological variability. Using the OpenProblems V2.0.0 batch integration benchmark, which combines 13 metrics into one overall score, our system discovered 40 novel methods that outperformed top expert-developed methods. The highest-scoring solution achieved a 14% overall improvement over the best published method (ComBat) by successfully combining two existing methods (ComBat and BBKNN).
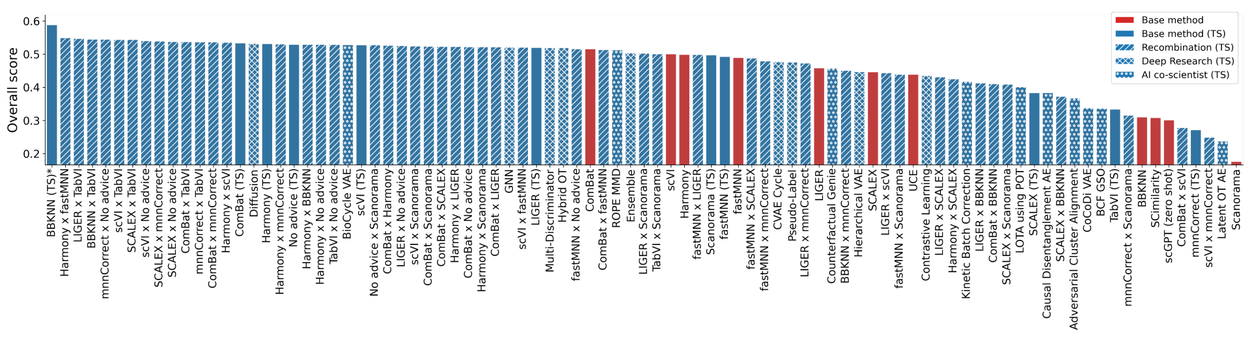
Overall leaderboard for OpenProblems benchmark v2.0.0 non-control methods. In blue are results from our system with and without recombination of ideas, and Gemini Deep Research. Click to enlarge image.
Public health: Prediction of U.S. COVID-19 hospitalizations
The primary U.S. benchmark for COVID-19 forecasting is the COVID-19 Forecast Hub (CovidHub), a large collaborative effort coordinated by the Centers for Disease Control and Prevention (CDC). CovidHub attracts competitive and methodologically diverse submissions from dozens of expert-led teams. Their task is to forecast new COVID-19 hospitalizations across all the U.S. states and its territories for up to a month ahead. These forecasts are evaluated using average weighted interval score (WIS), which assesses the quality of probabilistic forecasts by summarizing a model's performance across all locations for every weekly prediction over the season. Individual submissions are then aggregated into the CovidHub Ensemble model, which is considered the gold standard in the U.S. for forecasting COVID-19 hospitalizations. Our system generated 14 models that outperform the official CovidHub Ensemble.
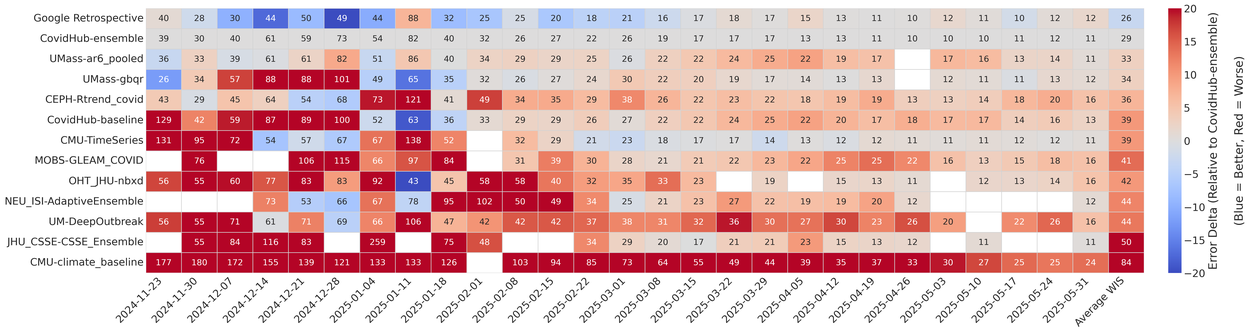
Time-series leaderboard showing weekly forecasting performance for teams participating in the COVID-19 Forecast Hub, ordered by absolute average WIS (number within each cell). Scores are aggregated across 52 jurisdictions and four forecast horizons. The cell’s background color visualizes the performance relative to the CovidHub-ensemble, with blue indicating a lower (better) WIS and red indicating a higher (worse) WIS. Our method, the top row of the table (Google Retrospective) outperforms CovidHub-ensemble. Click to enlarge image.
Geospatial analysis: Segmentation of remote sensing images
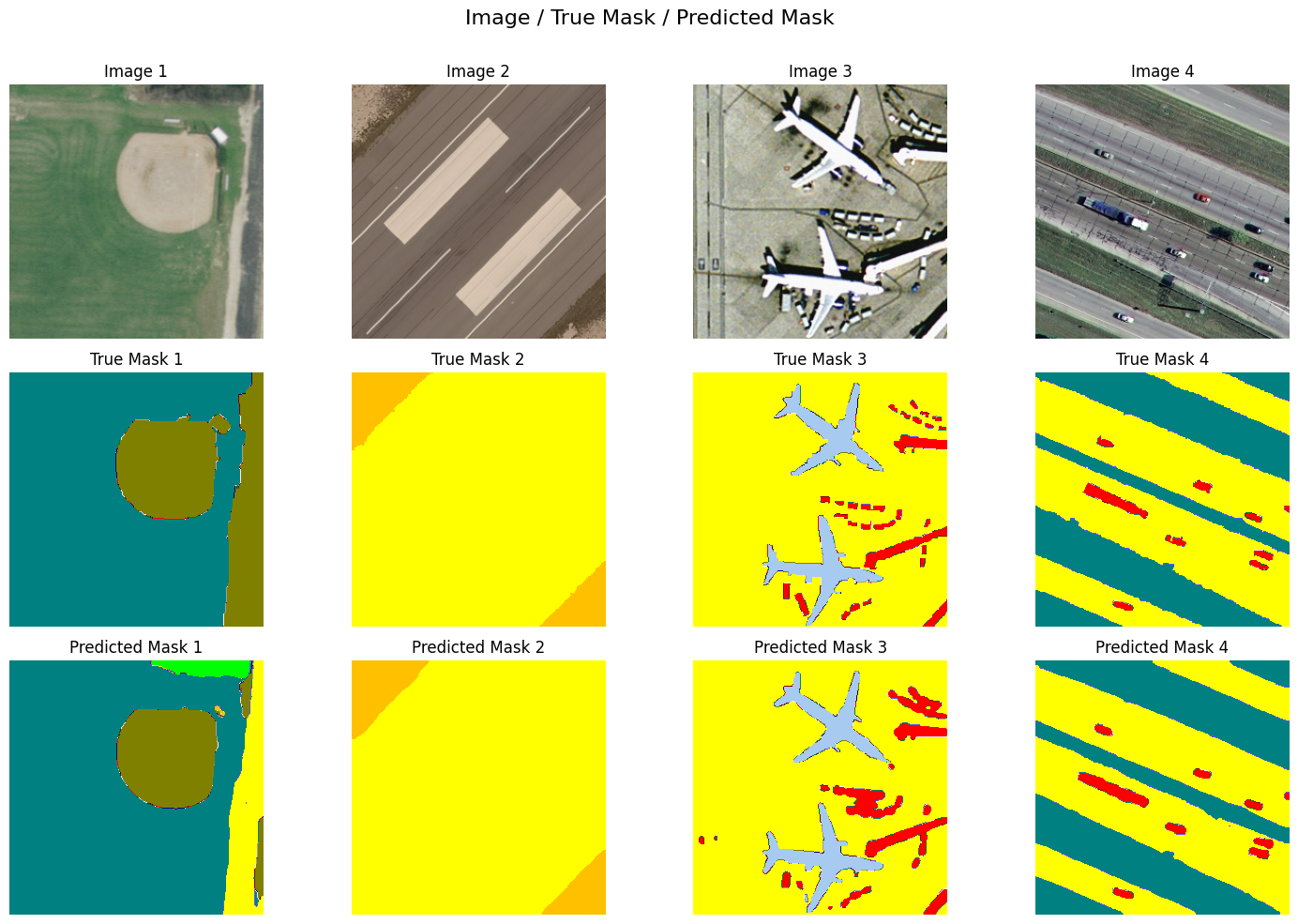
Semantic segmentation of high-resolution remote sensing images is a common problem in geospatial analysis, and is essential for diverse applications, ranging from monitoring land use, assessing the environmental impacts of human activity, and managing natural disasters. This task, which involves accurately assigning class labels to individual pixels in an image, requires a model to develop a spatial and contextual understanding of the scene, identifying not just what objects are present, but precisely where their boundaries lie.
Using the dense labeling remote sensing dataset (DLRSD) benchmark, which evaluates methods using a mean intersection over union (mIoU), the top three solutions generated by our system are slightly better than current state of the art, with mIoU greater than 0.80. All three solutions build upon existing models, libraries and strategies. Two leverage standard UNet++ and U-Net models but paired with powerful encoders pre-trained on ImageNet. The third uses SegFormer, a state of the art Transformer-based architecture. All three employ extensive test-time augmentation (TTA).
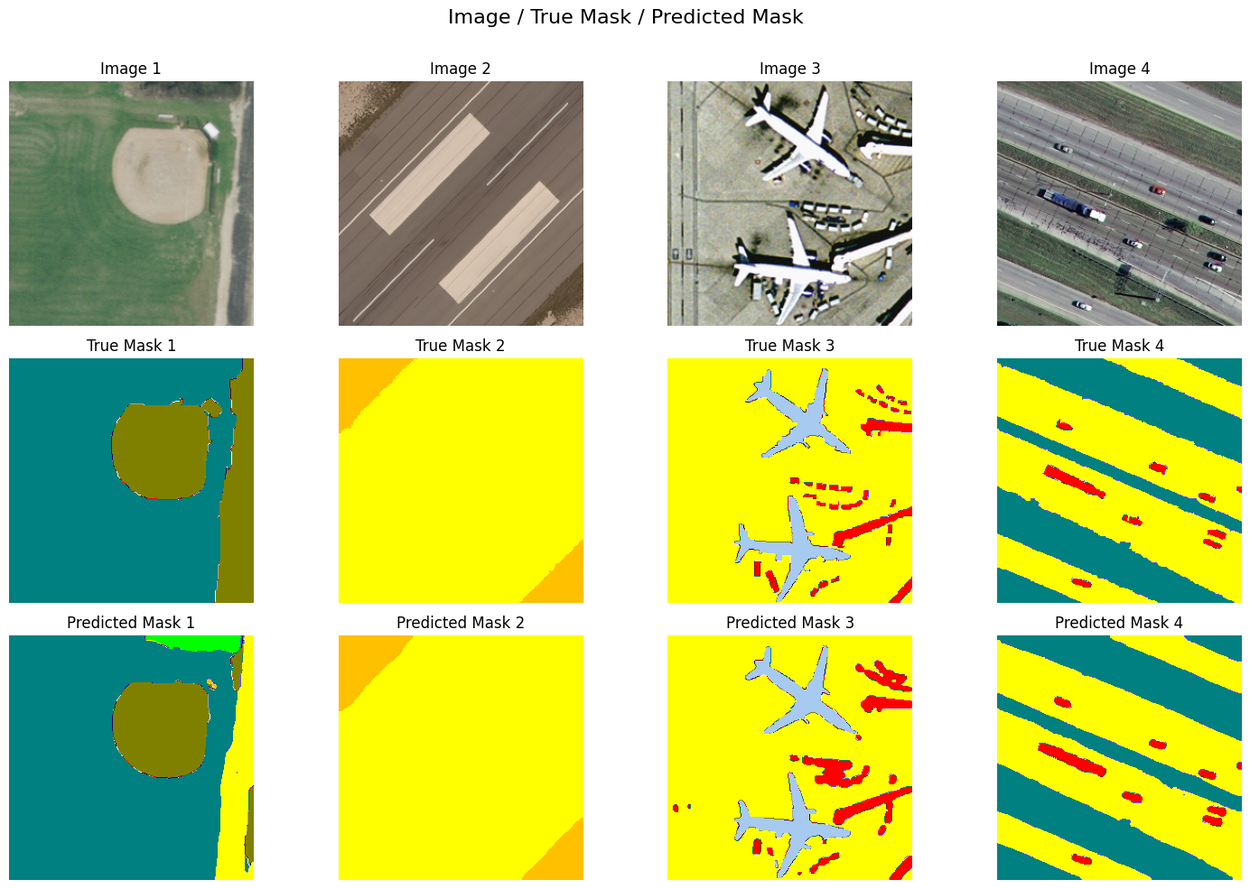
The input to remote sensing segmentation models is an image (top row), and the output is a new image, often called a segmentation mask, where each pixel is assigned a specific class label. The middle row is the true mask as provided by the DLRSD benchmark. The bottom row is segmentation masks generated using our system's top scoring solution. High-scoring segmentation models will have close visual similarity to the ground truth mask.
Neuroscience: Whole-brain neural activity prediction
We applied our method to the Zebrafish Activity Prediction Benchmark (ZAPBench), a recent benchmark for forecasting the activity of over 70,000 neurons across an entire vertebrate brain. Our system discovered a novel time-series forecasting model that achieved state-of-the-art performance, surpassing all existing baselines. This includes a computationally intensive, video-based model that forecasts 3D volumes and was the previous top performing solution. As a proof of concept, we also demonstrated that our system can design hybrid models that incorporate a biophysical neuron simulator (Jaxley), paving the way for more interpretable predictive models.
While each of these examples is compelling in its own right, our system to generate empirical software is striking in its generalizability. We additionally evaluated our system in the context of mathematics on the task of numerical evaluation of difficult integrals. In this task, our system generated a solution that correctly evaluated 17 out of 19 held-out integrals, where the standard numerical method failed. Lastly, we evaluated our system on the general problem of time series forecasting, using the General Time Series Forecasting Model Evaluation (GIFT-Eval), a benchmark derived from 28 datasets spanning seven diverse domains, with 10 different frequencies, from seconds to years. Our system successfully created a unified, general purpose forecasting library from scratch, by hill climbing with a single code on the average mean absolute scaled error on the entire GIFT-Eval dataset. See the paper for more details.
Conclusion
Recent advances in LLMs have already given researchers worldwide new ways to easily engage with knowledge and ideas, and LLMs are increasingly being pursued as a means of automating the rote and toilsome aspects of scientific research. We explored whether LLMs could be useful for the ubiquitous, essential, and highly time-consuming task of producing custom software for evaluating and iteratively improving scientific hypotheses, motivated by the possibility of a future where scientists can easily, rapidly, and systematically investigate hundreds or thousands of potential solutions to the questions and problems that motivate their research. Our system quickly generates expert-level solutions reducing the time required for exploration of a set of ideas from months to hours or days. This promises to save significant time for scientists, from students to professors, to focus on truly creative and critical challenges, and to continue to define and prioritize the fundamental research questions and societal challenges that scientific research can help address.
Acknowledgements
We thank and acknowledge the contributions from all of the co-authors of the manuscript. Thanks to Shibl Mourad, John Platt, Erica Brand, Katherine Chou, Ronit Levavi Morad, Yossi Matias, and James Manyika for their support and leadership.
-
Labels:
- General Science
- Generative AI