Accelerating scientific breakthroughs with an AI co-scientist
February 19, 2025
Juraj Gottweis, Google Fellow, and Vivek Natarajan, Research Lead
We introduce AI co-scientist, a multi-agent AI system built with Gemini 2.0 as a virtual scientific collaborator to help scientists generate novel hypotheses and research proposals, and to accelerate the clock speed of scientific and biomedical discoveries.
In the pursuit of scientific advances, researchers combine ingenuity and creativity with insight and expertise grounded in literature to generate novel and viable research directions and to guide the exploration that follows. In many fields, this presents a breadth and depth conundrum, since it is challenging to navigate the rapid growth in the rate of scientific publications while integrating insights from unfamiliar domains. Yet overcoming such challenges is critical, as evidenced by the many modern breakthroughs that have emerged from transdisciplinary endeavors. For example, Emmanuelle Charpentier and Jennifer Doudna won the 2020 Nobel Prize in Chemistry for their work on CRISPR, which combined expertise ranging from microbiology to genetics to molecular biology.
Motivated by unmet needs in the modern scientific discovery process and building on recent AI advances, including the ability to synthesize across complex subjects and to perform long-term planning and reasoning, we developed an AI co-scientist system. The AI co-scientist is a multi-agent AI system that is intended to function as a collaborative tool for scientists. Built on Gemini 2.0, AI co-scientist is designed to mirror the reasoning process underpinning the scientific method. Beyond standard literature review, summarization and “deep research” tools, the AI co-scientist system is intended to uncover new, original knowledge and to formulate demonstrably novel research hypotheses and proposals, building upon prior evidence and tailored to specific research objectives.
Empowering scientists and accelerating discoveries with the AI co-scientist
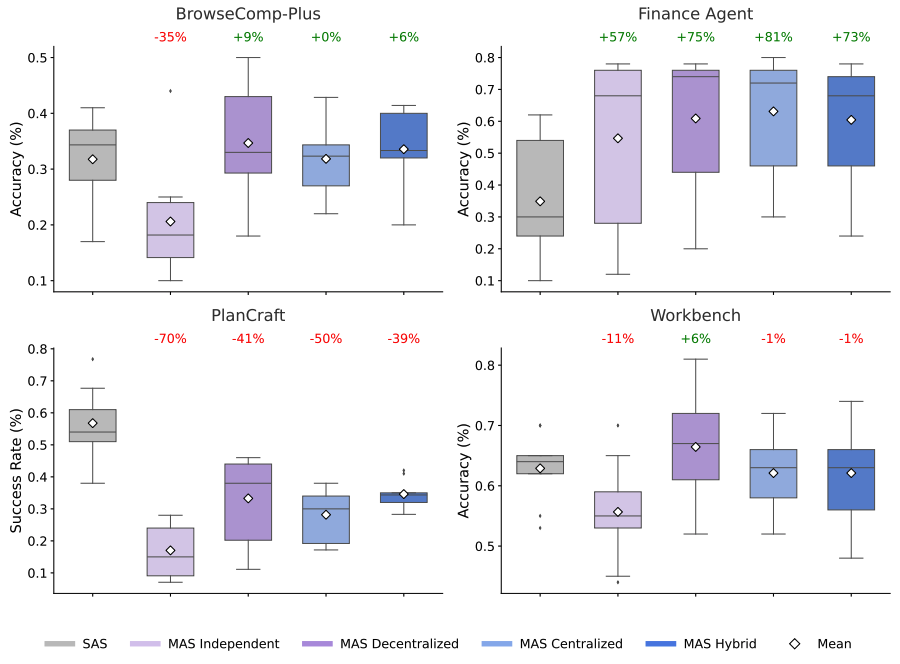
Given a scientist’s research goal that has been specified in natural language, the AI co-scientist is designed to generate novel research hypotheses, a detailed research overview, and experimental protocols. To do so, it uses a coalition of specialized agents — Generation, Reflection, Ranking, Evolution, Proximity and Meta-review — that are inspired by the scientific method itself. These agents use automated feedback to iteratively generate, evaluate, and refine hypotheses, resulting in a self-improving cycle of increasingly high-quality and novel outputs.
AI co-scientist overview.
Purpose-built for collaboration, scientists can interact with the system in many ways, including by directly providing their own seed ideas for exploration or by providing feedback on generated outputs in natural language. The AI co-scientist also uses tools, like web-search and specialized AI models, to enhance the grounding and quality of generated hypotheses.
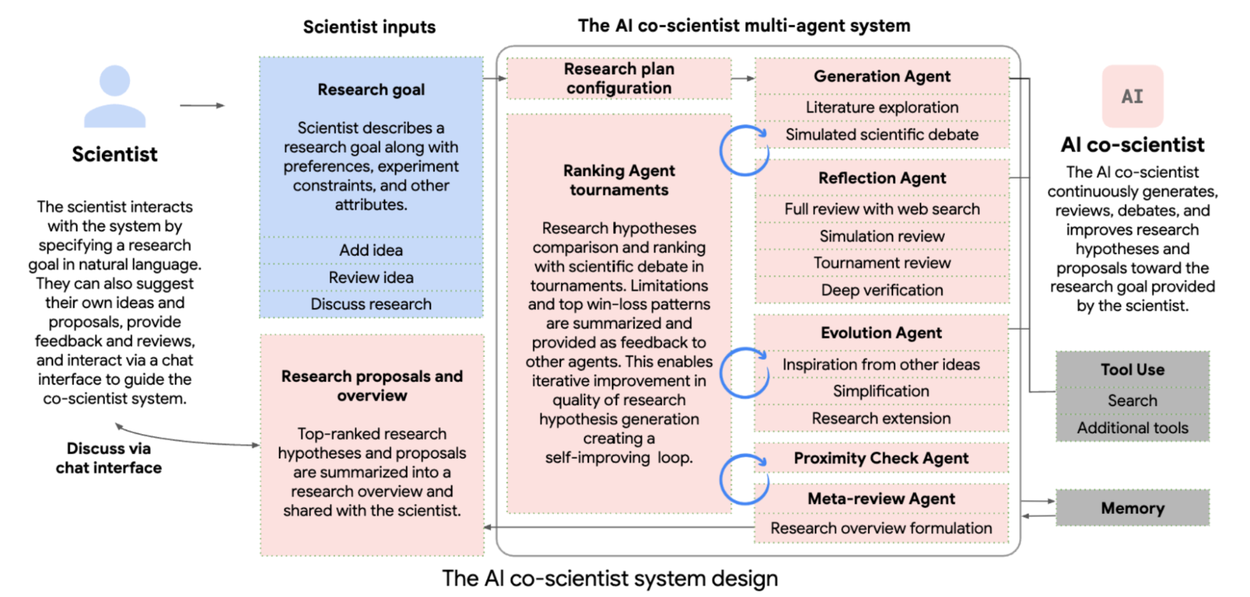
Illustration of the different components in the AI co-scientist multi-agent system and the interaction paradigm between the system and the scientist.
The AI co-scientist parses the assigned goal into a research plan configuration, managed by a Supervisor agent. The Supervisor agent assigns the specialized agents to the worker queue and allocates resources. This design enables the system to flexibly scale compute and to iteratively improve its scientific reasoning towards the specified research goal.
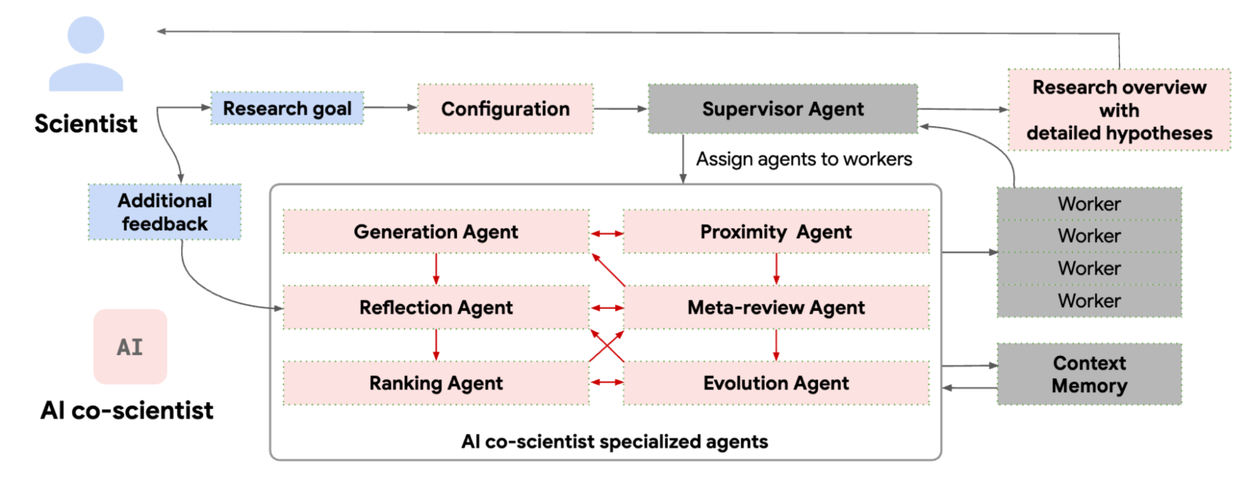
AI co-scientist system overview. Specialized agents (red boxes, with unique roles and logic); scientist input and feedback (blue boxes); system information flow (dark gray arrows); inter-agent feedback (red arrows within the agent section).
Scaling test-time compute for advanced scientific reasoning
The AI co-scientist leverages test-time compute scaling to iteratively reason, evolve, and improve outputs. Key reasoning steps include self-play–based scientific debate for novel hypothesis generation, ranking tournaments for hypothesis comparison, and an "evolution" process for quality improvement. The system's agentic nature facilitates recursive self-critique, including tool use for feedback to refine hypotheses and proposals.
The system's self-improvement relies on the Elo auto-evaluation metric derived from its tournaments. Due to their core role, we assessed whether higher Elo ratings correlate with higher output quality. We analyzed the concordance between Elo auto-ratings and GPQA benchmark accuracy on its diamond set of challenging questions, and we found that higher Elo ratings positively correlate with a higher probability of correct answers.
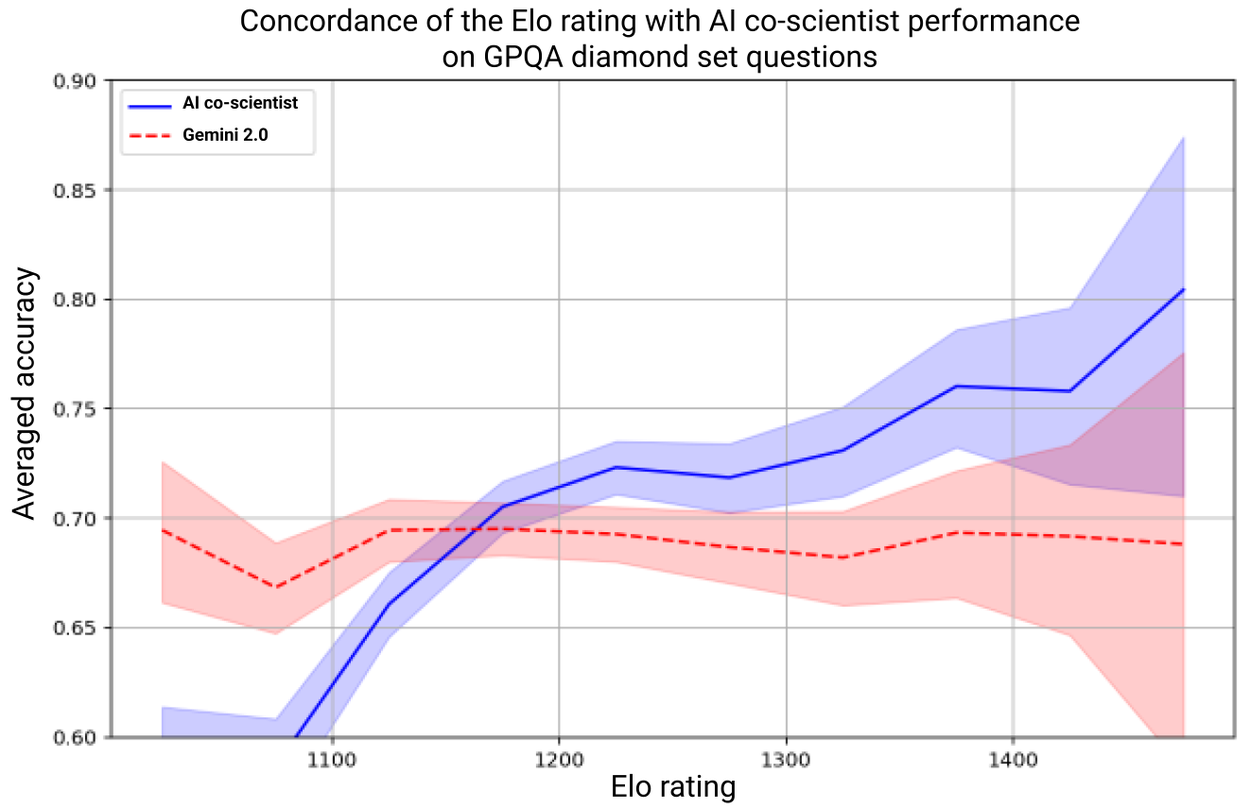
Average accuracy of the AI co-scientist (blue line) and reference Gemini 2.0 (red line) responses on GPQA diamond questions, grouped by Elo rating. The Elo is an auto-evaluation and is not based on an independent ground truth.
Seven domain experts curated 15 open research goals and best guess solutions in their field of expertise. Using the automated Elo metric we observed that the AI co-scientist outperformed other state-of-the-art agentic and reasoning models for these complex problems. The analysis reproduced the benefits of scaling test-time compute using inductive biases derived from the scientific method. As the system spends more time reasoning and improving, the self-rated quality of results improve and surpass models and unassisted human experts.
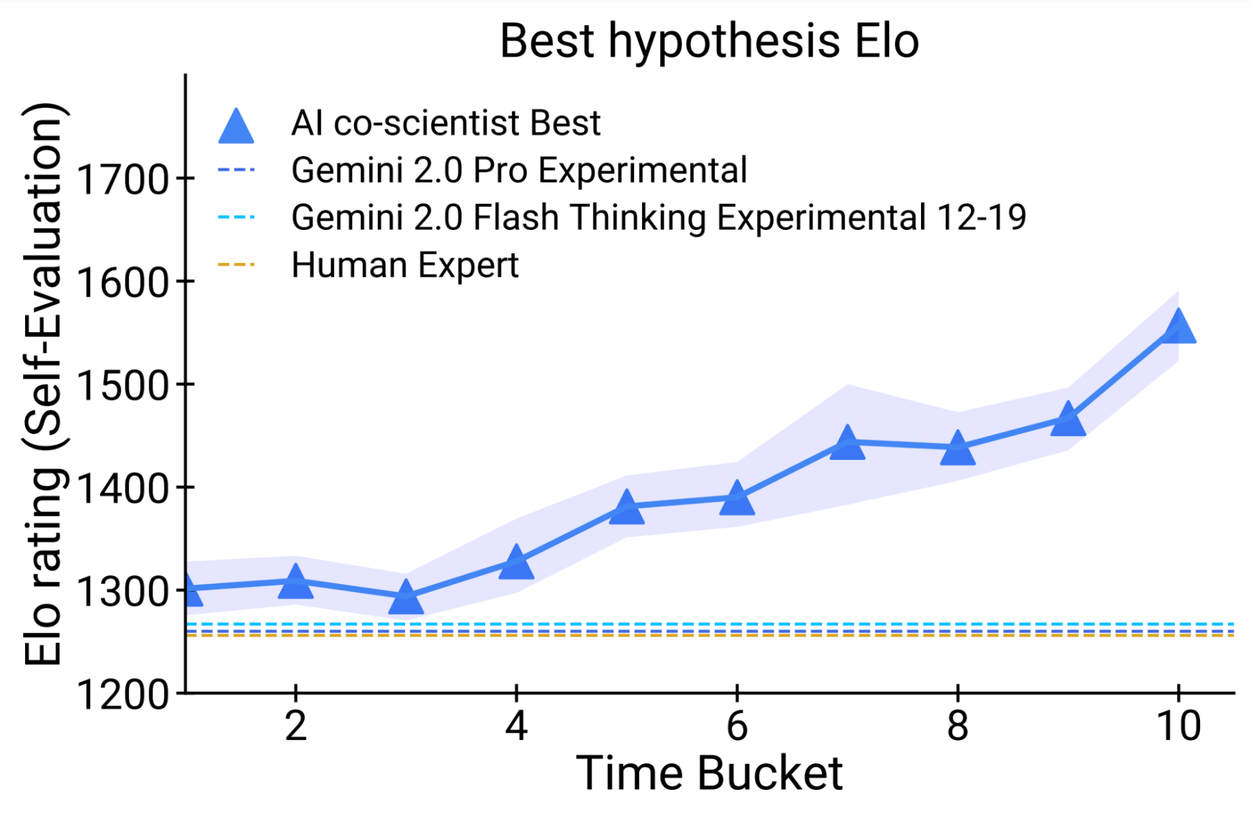
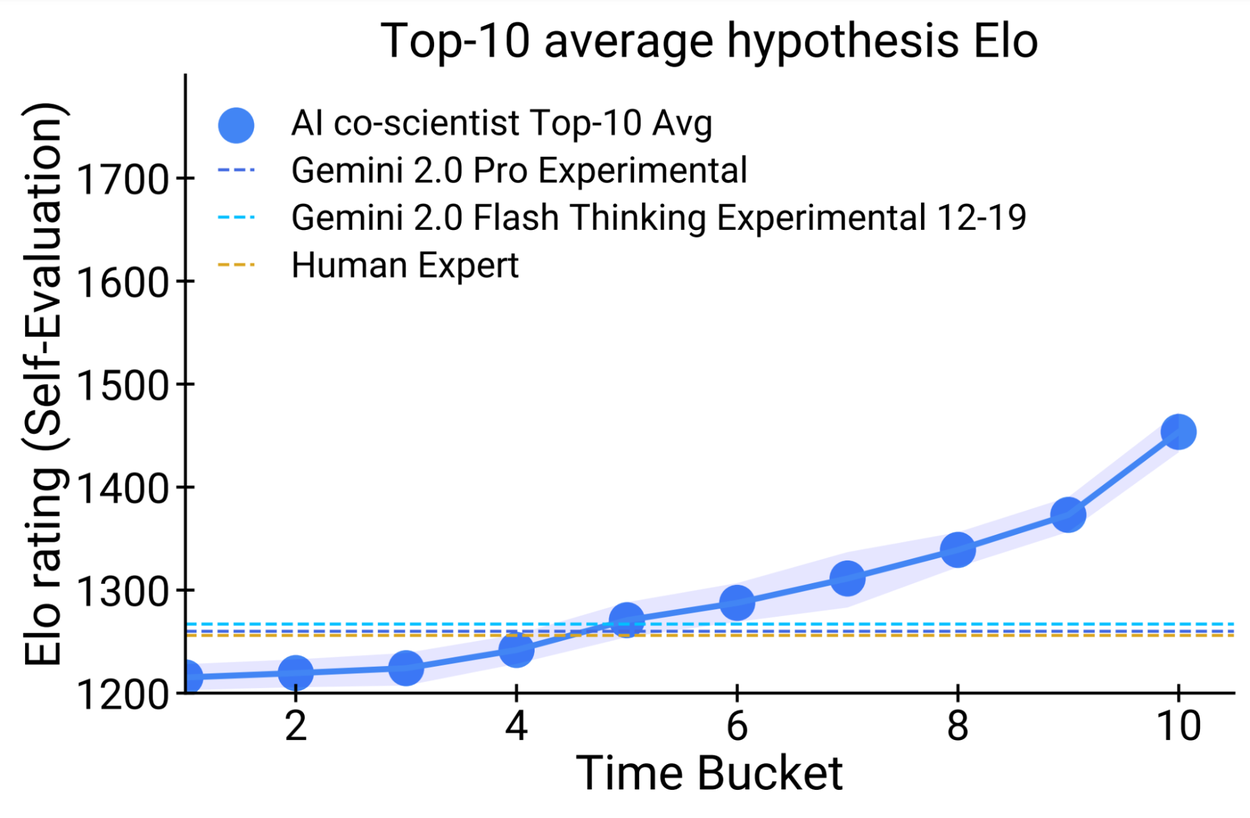
Performance of the AI co-scientist improves as the system spends more time in computation. This can be seen in the automated Elo metric gradually improving over other baselines. Top: Elo progression of the best rated hypothesis. Bottom: Elo progression of the average of top-10 hypotheses.
On a smaller subset of 11 research goals, experts assessed the novelty and impact of the AI co-scientist–generated results compared to other relevant baselines; they also provided overall preference. While the sample size was small, experts assessed the AI co-scientist to have higher potential for novelty and impact, and preferred its outputs compared to other models. Further, these human expert preferences also appeared to be concordant with the previously introduced Elo auto-evaluation metric.
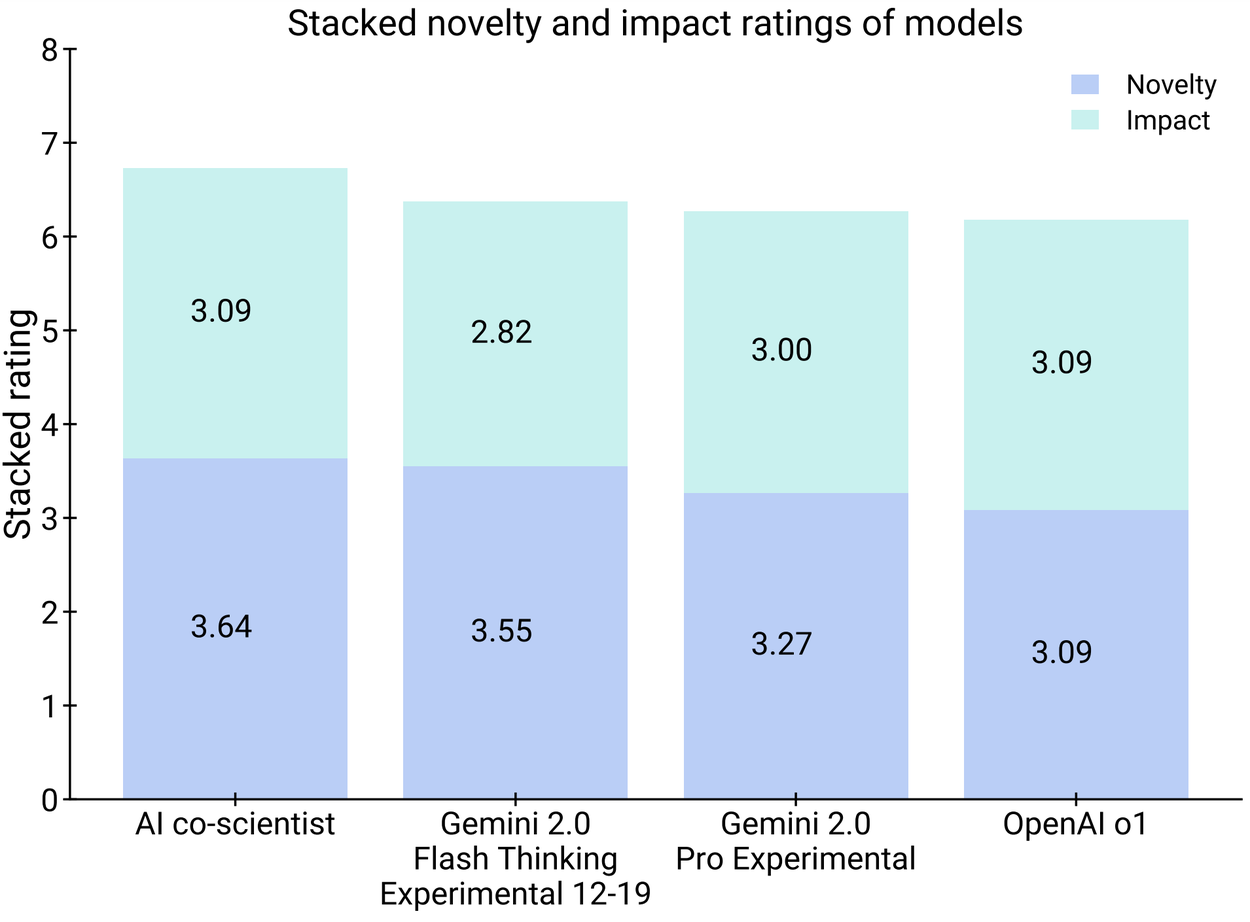
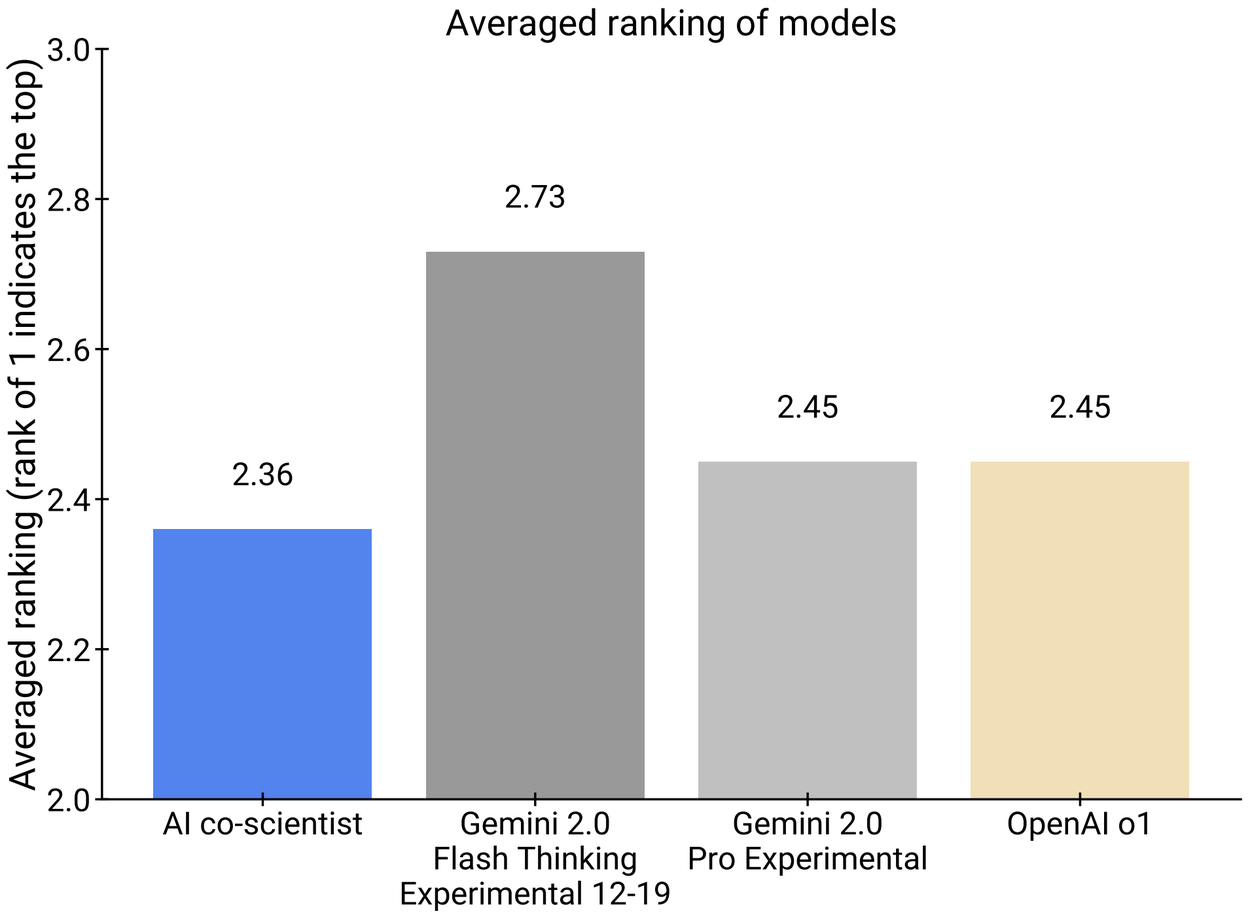
Human experts assessed the AI co-scientist results to have higher potential for novelty and impact (left) and preferred it compared to other models (right).
Validation of novel AI co-scientist hypotheses with real-world laboratory experiments
To assess the practical utility of the system’s novel predictions, we evaluated end-to-end laboratory experiments probing the AI co-scientist–generated hypotheses and research proposals in three key biomedical applications: drug repurposing, proposing novel treatment targets, and elucidating the mechanisms underlying antimicrobial resistance. These settings all involved expert-in-the-loop guidance and spanned an array of complexities:
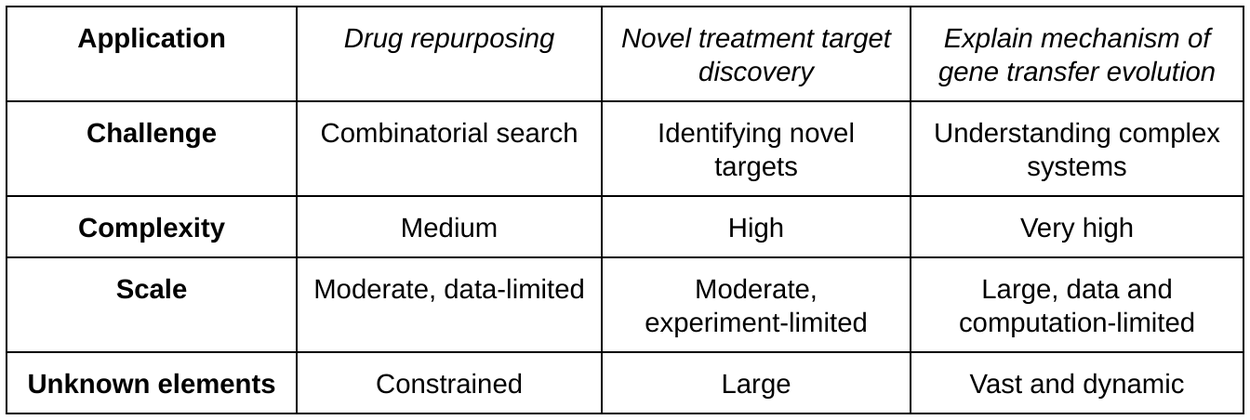
Drug repurposing for acute myeloid leukaemia
Drug development is an increasingly time-consuming and expensive process in which new therapeutics require many aspects of the discovery and development process to be restarted for each indication or disease. Drug repurposing addresses this challenge by discovering new therapeutic applications for existing drugs beyond their original intended use. But, due to the complexity of the task, it demands extensive interdisciplinary expertise.
We applied the AI co-scientist to assist with the prediction of drug repurposing opportunities and, with our partners, validated predictions through computational biology, expert clinician feedback, and in vitro experiments.
Notably, the AI co-scientist proposed novel repurposing candidates for acute myeloid leukemia (AML). Subsequent experiments validated these proposals, confirming that the suggested drugs inhibit tumor viability at clinically relevant concentrations in multiple AML cell lines.
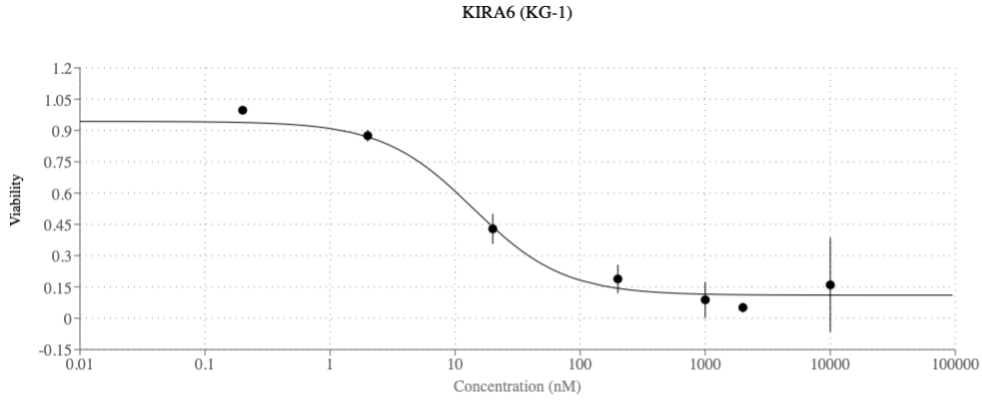
Dose-response curves of one of the three novel AI co-scientist–predicted AML repurposing drugs. KIRA6 inhibits KG-1 (AML cell line) viability at clinically relevant concentrations. Being able to reduce cancer cell viability at lower drug concentrations is advantageous for multiple reasons, e.g., as it reduces the potential for off-target side effects.
Advancing target discovery for liver fibrosis
Identifying novel treatment targets is more complex than drug repurposing, and often leads to inefficient hypothesis selection and poor prioritization for in vitro and in vivo experiments. AI-assisted target discovery helps to streamline the process of experimental validation, potentially helping to reduce development time costs.
We probed the AI co-scientist system's ability to propose, rank, and generate hypotheses and experimental protocols for target discovery hypotheses, focusing on liver fibrosis. The AI co-scientist demonstrated its potential by identifying epigenetic targets grounded in preclinical evidence with significant anti-fibrotic activity in human hepatic organoids (3D, multicellular tissue cultures derived from human cells and designed to mimic the structure and function of the human liver). These findings will be detailed in an upcoming report led by collaborators at Stanford University.
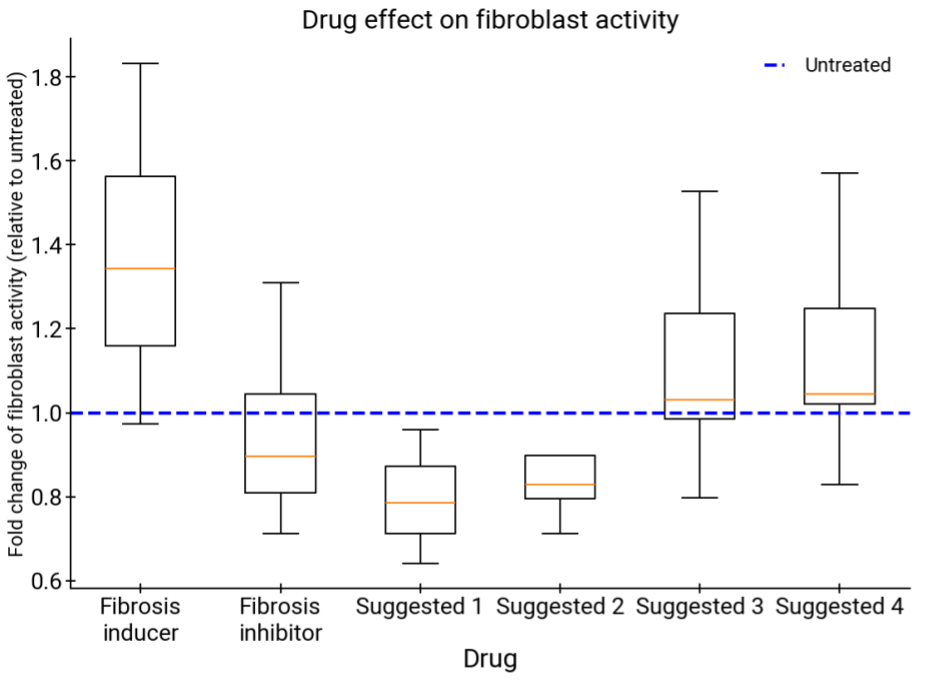
Comparison of treatments derived from AI co-scientist–suggested liver fibrosis targets versus a fibrosis inducer (negative control) and an inhibitor (positive control). All treatments suggested by AI co-scientist show promising activity (p-values for all suggested drugs are <0.01), including candidates that possibly reverse a disease phenotype. Results are detailed in an upcoming report from our Stanford University collaborators.
Explaining mechanisms of antimicrobial resistance
As a third validation, we focused on generating hypotheses to explain bacterial gene transfer evolution mechanisms related to antimicrobial resistance (AMR) — microbes' evolved mechanisms to resist infection-treating drugs. This is another complex challenge that involves understanding the molecular mechanisms of gene transfer (conjugation, transduction, and transformation) alongside the ecological and evolutionary pressures that drive AMR genes to spread.
For this test, expert researchers instructed the AI co-scientist to explore a topic that had already been subject to novel discovery in their group, but had not yet been revealed in the public domain, namely, to explain how capsid-forming phage-inducible chromosomal islands (cf-PICIs) exist across multiple bacterial species. The AI co-scientist system independently proposed that cf-PICIs interact with diverse phage tails to expand their host range. This in silico discovery, which had been experimentally validated in the original novel laboratory experiments performed prior to use of the AI co-scientist system, are described in co-timed manuscripts (1, 2) with our collaborators at the Fleming Initiative and Imperial College London. This illustrates the value of the AI co-scientist system as an assistive technology, as it was able to leverage decades of research comprising all prior open access literature on this topic.
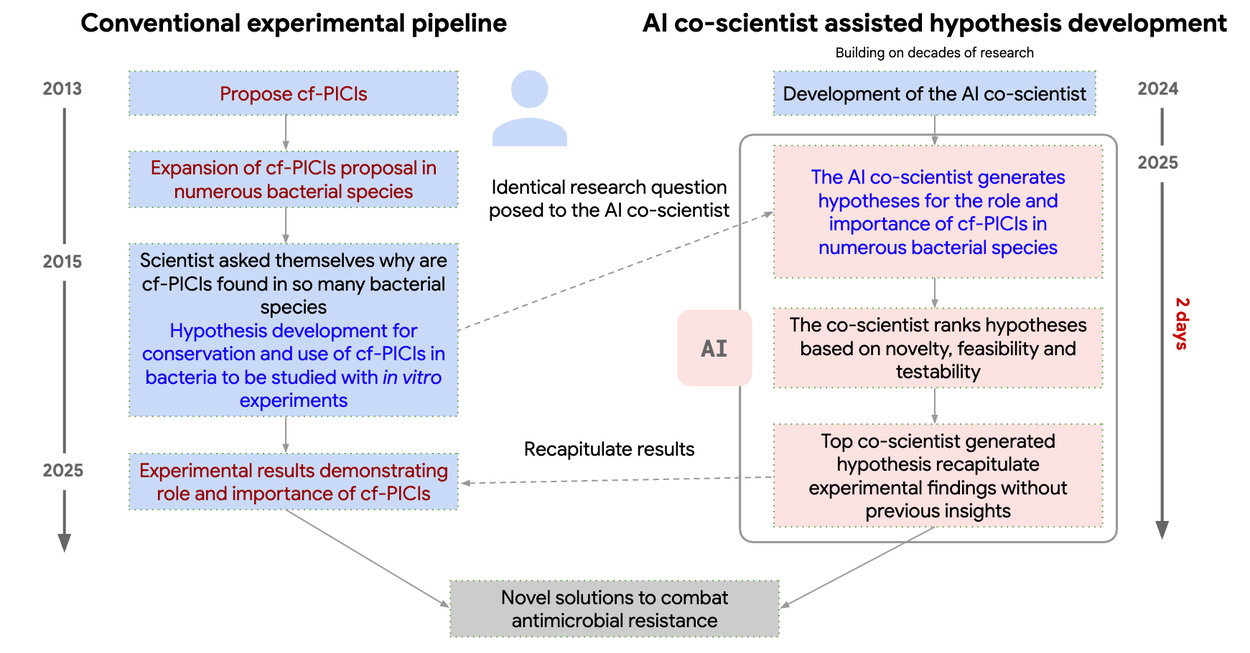
Timeline of AI co-scientist re-discovery of a novel gene transfer mechanism. Blue: Experimental research pipeline timeline for cf-PICI mobilization discovery. Red: AI co-scientist development and recapitulation of these key findings (without prior knowledge).
Limitations and outlook
In our report we address several limitations of the system and opportunities for improvement, including enhanced literature reviews, factuality checking, cross-checks with external tools, auto-evaluation techniques, and larger-scale evaluation involving more subject matter experts with varied research goals. The AI co-scientist represents a promising advance toward AI-assisted technologies for scientists to help accelerate discovery. Its ability to generate novel, testable hypotheses across diverse scientific and biomedical domains — some already validated experimentally — and its capacity for recursive self-improvement with increased compute, demonstrate its potential to accelerate scientists' efforts to address grand challenges in science and medicine. We look forward to responsible exploration of the potential of the AI co-scientist as an assistive tool for scientists. This project illustrates how collaborative and human-centred AI systems might be able to augment human ingenuity and accelerate scientific discovery.
Announcing Trusted Tester access to the AI co-scientist system
We are excited by the early promise of the AI co-scientist system and believe it is important to evaluate its strengths and limitations in science and biomedicine more broadly. To facilitate this responsibly we will be enabling access to the system for research organizations through a Trusted Tester Program. We encourage interested research organizations around the world to consider joining this program here.
Acknowledgements
The research described here is a joint effort between many Google Research, Google Deepmind and Google Cloud AI teams. We thank our co-authors at Fleming Initiative and Imperial College London, Houston Methodist Hospital, Sequome, and Stanford University — José R Penadés, Tiago R D Costa, Vikram Dhillon, Eeshit Dhaval Vaishnav, Byron Lee, Jacob Blum and Gary Peltz. We appreciate Subhashini Venugopalan and Yun Liu for their detailed feedback on the manuscripts described here. We are also grateful to the many incredible scientists across institutions providing detailed technical and expert feedback — please refer to our report to see the voices and minds that aided this work. We also thank our teammates Resham Parikh, Taylor Goddu, Siyi Kou, Rachelle Sico, Amanda Ferber, Cat Kozlowski, Alison Lentz, KK Walker, Roma Ruparel, Jenn Sturgeon, Lauren Winer, Juanita Bawagan, Tori Milner, MK Blake, Kalyan Pamarthy for their support. Finally, we also thank John Platt, Michael Brenner, Zoubin Ghahramani, Dale Webster, Joelle Barral, Michael Howell, Susan Thomas, Jason Freidenfelds, Karen DeSalvo, Vladimir Vuskovic, Greg Corrado, Ronit Levavi Morad, Ali Eslami, Anna Koivuniemi, Royal Hansen, Andy Berndt, Noam Shazeer, Oriol Vinyals, Burak Gokturk, Amin Vahdat, Katherine Chou, Avinatan Hassidim, Koray Kavukcuoglu, Pushmeet Kohli, Yossi Matias, James Manyika, Jeff Dean and Demis Hassabis for their support.